7DIG
 
 | |
7E5J
 
 | | Crystal structure of beta-glucosidase from Thermoanaerobacterium saccharolyticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Molecules, 27, 2022
|
|
7WKR
 
 | |
4ES3
 
 | |
4ES1
 
 | |
4F3M
 
 | | Crystal structure of CRISPR-associated protein | | Descriptor: | 1,2-ETHANEDIOL, BH0337 protein, SULFATE ION | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-05-09 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System.
Structure, 20, 2012
|
|
7WUC
 
 | |
4ES2
 
 | |
3S5U
 
 | | Crystal structure of CRISPR associated protein | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity.
J. Biol. Chem., 286, 2011
|
|
3CMJ
 
 | |
4HZ6
 
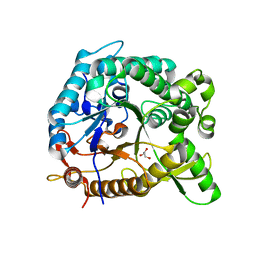 | | crystal structure of BglB | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the substrate recognition properties of beta-glucosidase.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4HZ8
 
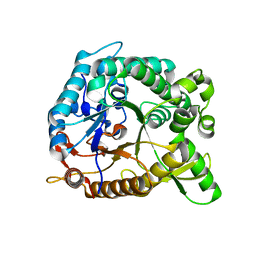 | | Crystal structure of BglB with natural substrate | | Descriptor: | Beta-glucosidase, beta-D-glucopyranose | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural insights into the substrate recognition properties of beta-glucosidase.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4HZ7
 
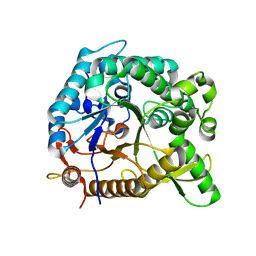 | | Crystal structure of BglB with glucose | | Descriptor: | beta-D-glucopyranose, beta-glucosidase | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate recognition properties of beta-glucosidase.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4H7A
 
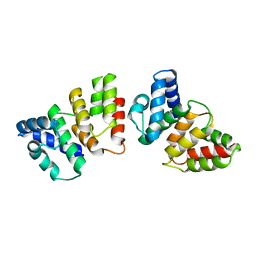 | | Crystal structure of CasB from Thermus thermophilus | | Descriptor: | CRISPR-associated protein Cse2 | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures.
Febs Lett., 586, 2012
|
|
4H79
 
 | | Crystal structure of CasB from Thermobifida fusca | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated protein, Cse2 family | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures.
Febs Lett., 586, 2012
|
|
5HZU
 
 | | Crystal structure of Dronpa-Ni2+ | | Descriptor: | Fluorescent protein Dronpa, NICKEL (II) ION | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZS
 
 | | Crystal structure of Dronpa-Co2+ | | Descriptor: | COBALT (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
8H8T
 
 | |
8H8V
 
 | |
8H8W
 
 | |
8H8U
 
 | |
5Z6V
 
 | |
8H2A
 
 | |
8H2B
 
 | |
6K1G
 
 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
