8SHH
 
 | | Crystal structure of EvdS6 decarboxylase in ligand free state | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8VKR
 
 | | CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VID
 
 | | CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VIB
 
 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VKQ
 
 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
6EC3
 
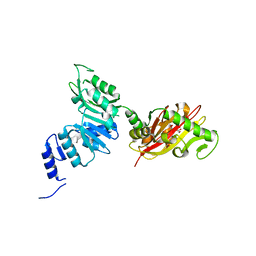 | | Crystal Structure of EvdMO1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyltransferase domain-containing protein, NICKEL (II) ION | | Authors: | McCulloch, K.M, Iverson, T.M, Starbird, C.A, Perry, N.A, Chen, Q, Berndt, S, Yamakawa, I, Loukachevitch, L.V. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
3OT9
 
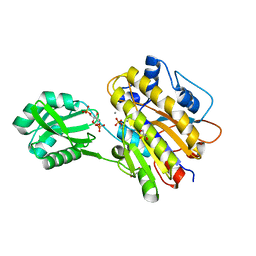 | | Phosphopentomutase from Bacillus cereus bound to glucose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Phalen, V, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-09-10 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3M8Z
 
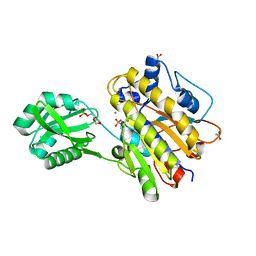 | | Phosphopentomutase from Bacillus cereus bound with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3MXL
 
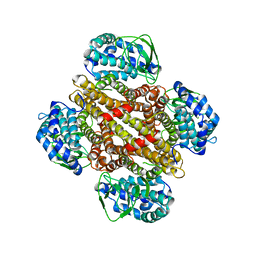 | |
4LRE
 
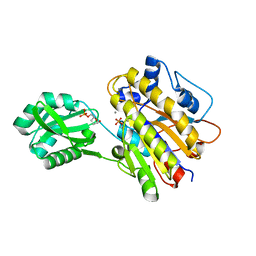 | | Phosphopentomutase soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LRA
 
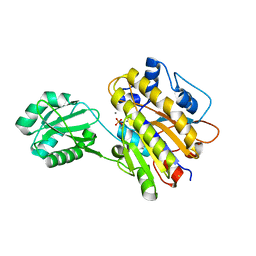 | | Phosphopentomutase S154G variant | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LRD
 
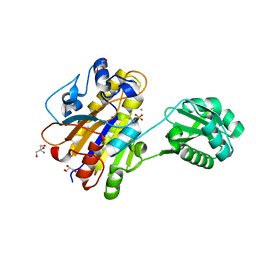 | | Phosphopentomutase 4H11 variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LRC
 
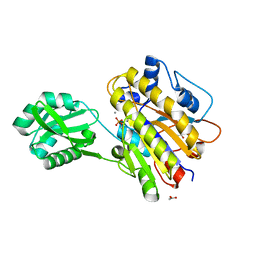 | | Phosphopentomutase V158L variant | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LR7
 
 | | Phosphopentomutase S154A variant | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LR8
 
 | | Phosphopentomutase S154A variant soaked with ribose 5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LRB
 
 | | Phosphopentomutase S154G variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, GLYCEROL, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4MBO
 
 | | 1.65 Angstrom Crystal Structure of Serine-rich Repeat Adhesion Glycoprotein (Srr1) from Streptococcus agalactiae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4LR9
 
 | | Phosphopentomutase S154A variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4MBR
 
 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | Descriptor: | Serine-rich repeat protein 2 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4LRF
 
 | | Phosphopentomutase S154G variant soaked with ribose 5-phosphate | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
1MJG
 
 | | CRYSTAL STRUCTURE OF BIFUNCTIONAL CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE(CODH/ACS) FROM MOORELLA THERMOACETICA (F. CLOSTRIDIUM THERMOACETICUM) | | Descriptor: | ACETATE ION, CARBON MONOXIDE DEHYDROGENASE BETA SUBUNIT, COPPER (I) ION, ... | | Authors: | Doukov, T.I, Iverson, T.M, Seravalli, J, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2002-08-27 | | Release date: | 2003-01-28 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA
synthase
Science, 298, 2002
|
|
3QC6
 
 | | GspB | | Descriptor: | CALCIUM ION, GLYCEROL, NITROGEN MOLECULE, ... | | Authors: | Pyburn, T.M. | | Deposit date: | 2011-01-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors.
Plos Pathog., 7, 2011
|
|
8FTJ
 
 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | Authors: | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | Deposit date: | 2023-01-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
