1W0N
 
 | | Structure of uncomplexed Carbohydrate Binding Domain CBM36 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, MAGNESIUM ION, ... | | Authors: | Jamal, S, Boraston, A.B, Davies, G.J. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1UX7
 
 | | Carbohydrate-Binding Module CBM36 in complex with calcium and xylotriose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, SULFATE ION, ... | | Authors: | Davies, G.J, Boraston, A.B, Jamal, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1UY4
 
 | |
1UY3
 
 | |
1UY1
 
 | |
1UY2
 
 | |
3QSP
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 non-productive substrate complex with alpha-1,6-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3REN
 
 | | CPF_2247, a novel alpha-amylase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase, family 8, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Boraston, A.B. | | Deposit date: | 2011-04-04 | | Release date: | 2011-05-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of CPF_2247, a novel alpha-amylase from Clostridium perfringens.
Proteins, 79, 2011
|
|
3QT9
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Clostridium perfringens CPE0426 complexed with alpha-1,6-linked 1-thio-alpha-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426, alpha-D-mannopyranose-(1-6)-6-thio-alpha-D-mannopyranose | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3QT3
 
 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426 | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3QRY
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 1-deoxymannojirimycin complex | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-18 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
7JNB
 
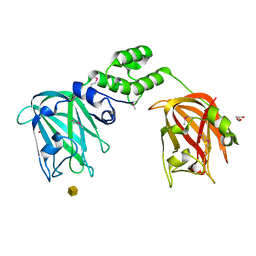 | |
7JS4
 
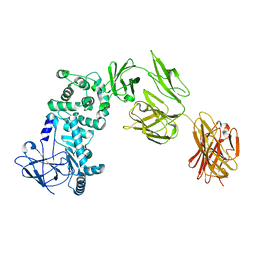 | |
7JWF
 
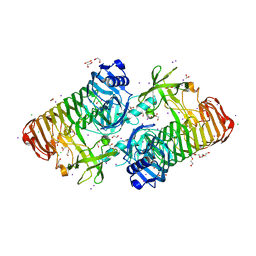 | | Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2020-08-25 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structure of a family 110 glycoside hydrolase provides insight into the hydrolysis of alpha-1,3-galactosidic linkages in lambda-carrageenan and blood group antigens.
J.Biol.Chem., 295, 2020
|
|
7JRM
 
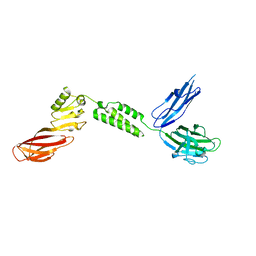 | |
7JND
 
 | |
7JNF
 
 | |
7JFS
 
 | |
7JRL
 
 | | The structure of CBM51-2 in complex with GlcNAc and INT domains from Clostridium perfringens ZmpB | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-08-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecturally complex O -glycopeptidases are customized for mucin recognition and hydrolysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JW4
 
 | | Crystal structure of PdGH110B in complex with D-galactose | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 110, NICKEL (II) ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | The structure of a family 110 glycoside hydrolase provides insight into the hydrolysis of alpha-1,3-galactosidic linkages in lambda-carrageenan and blood group antigens.
J.Biol.Chem., 295, 2020
|
|
7JTV
 
 | |
5A6J
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A6K
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-NGT | | Descriptor: | (2S,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, GH20C | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Second Beta-Hexosaminidase Encoded in the Streptococcus Pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
J.Biol.Chem., 290, 2015
|
|
5AMU
 
 | | IglE I39A,Y40A,V44A | | Descriptor: | IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure, Dimerisation and Impact on Intramacrophage Replication of Igle in Francisella Novicida
To be Published
|
|
5KD2
 
 | |
