5ZPL
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 7 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2ZPB
 
 | | nitrosylated Fe-type nitrile hydratase | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-09 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPI
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 440min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
5ZP0
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 8 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPF
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 288 K (3) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPT
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 10 at 288 K (2) | | Descriptor: | COPPER (II) ION, PHENYLACETALDEHYDE, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
2YN9
 
 | | Cryo-EM structure of gastric H+,K+-ATPase with bound rubidium | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Friedrich, T, Fujiyoshi, Y. | | Deposit date: | 2012-10-13 | | Release date: | 2012-11-07 | | Last modified: | 2014-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Cryo-Em Structure of Gastric H+,K+-ATPase with a Single Occupied Cation-Binding Site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ZPG
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 120min at 293K | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2Z9V
 
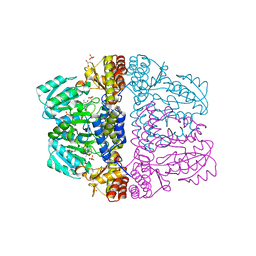 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZQ9
 
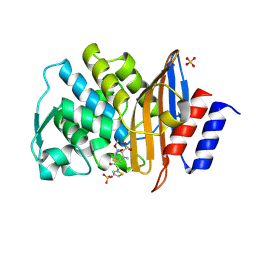 | | Cephalothin acyl-intermediate structure of class a beta-lactamase Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase Toho-1, CEPHALOTHIN, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
7F9O
 
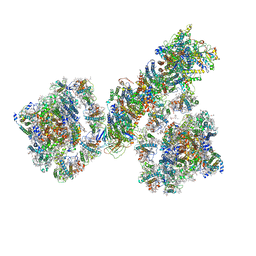 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EW6
 
 | | Barley photosystem I-LHCI-Lhca5 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-24 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EU3
 
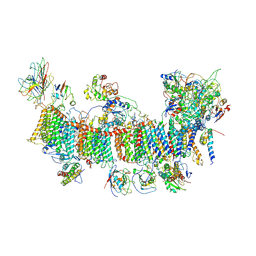 | | Chloroplast NDH complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EWK
 
 | | Barley photosystem I-LHCI-Lhca6 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7CBD
 
 | | Catalytic domain of Cellulomonas fimi Cel6B | | Descriptor: | Exoglucanase A | | Authors: | Nakamura, A, Ishiwata, D, Visootsat, A, Uchiyama, T, Mizutani, K, Kaneko, S, Murata, T, Igarashi, K, Iino, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.
J.Biol.Chem., 295, 2020
|
|
2YX9
 
 | | Crystal structure of D298K copper amine oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Trapping of a dopaquinone intermediate in the TPQ cofactor biogenesis in a copper-containing amine oxidase from Arthrobacter globiformis.
J.Am.Chem.Soc., 129, 2007
|
|
2ZFH
 
 | |
3WE2
 
 | |
3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
2ZPY
 
 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
3WE3
 
 | |
3X3Y
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | Descriptor: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X42
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X41
 
 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
