8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8I
 
 | | De novo design protein -N14 | | Descriptor: | De novo design protein -N14 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8F
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7Z
 
 | | De novo design protein -N1 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7M
 
 | | De novo design protein -T01 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K83
 
 | | De novo design protein -N2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, De novo design protein, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K84
 
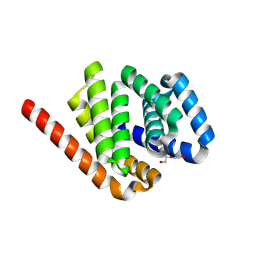 | | De novo design protein -N3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KAC
 
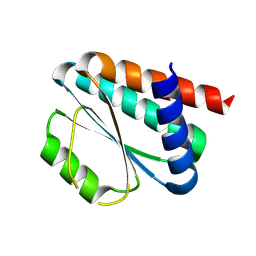 | | De novo design protein -NX1 | | Descriptor: | De novo design protein -NX1 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8G
 
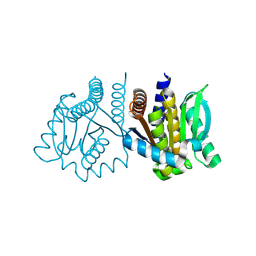 | | De novo design protein -N9 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC4
 
 | | De novo design protein -NA05 | | Descriptor: | De novo design protein -NA05 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KDQ
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein -T03 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC1
 
 | | De novo design protein -NX5 | | Descriptor: | De novo design protein -NX5 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC8
 
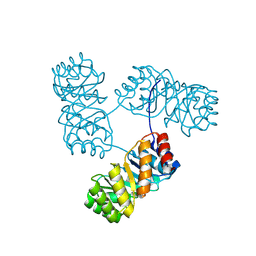 | | De novo design protein -T11 | | Descriptor: | De novo design protein -T11 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KCJ
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein -N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KA7
 
 | | De novo design protein -NB7 | | Descriptor: | De novo design protein -NB7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC5
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KA6
 
 | | De novo design protein -NA7 | | Descriptor: | De novo design protein -NA7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KCK
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein -N9 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC0
 
 | | De novo design protein -NB8 | | Descriptor: | De novo design protein -NB8 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8ZVX
 
 | | Crystal structure of snFPITE-n2 | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Chen, X, Yang, H. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A facultative plasminogen-independent thrombolytic enzyme from Sipunculus nudus.
Nat Commun, 16, 2025
|
|
8ZVS
 
 | | Crystal structure of snFPITE-n1 | | Descriptor: | SULFATE ION, phenylmethanesulfonic acid, snFPITE-n1 | | Authors: | Chen, X, Yang, H, Hu, Y.L. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | A facultative plasminogen-independent thrombolytic enzyme from Sipunculus nudus.
Nat Commun, 16, 2025
|
|
1MAE
 
 | |
1MAF
 
 | |
4I2R
 
 | | 2.15 Angstroms X-ray crystal structure of NAD- and alternative substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I1W
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
