2W5F
 
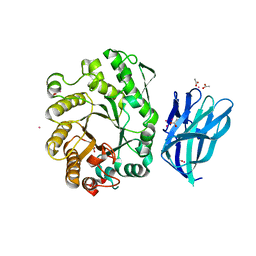 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2008-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WZE
 
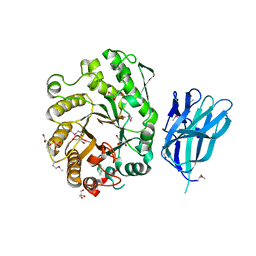 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-27 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WYS
 
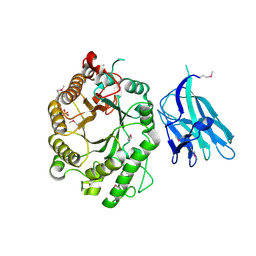 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, PHOSPHATE ION, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-20 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
1DGJ
 
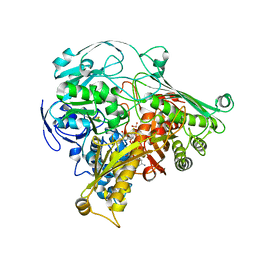 | | CRYSTAL STRUCTURE OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ALDEHYDE OXIDOREDUCTASE, FE2/S2 (INORGANIC) CLUSTER, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Rebelo, J.M, Macieira, S, Dias, J.M, Huber, R, Romao, M.J. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774.
J.Mol.Biol., 297, 2000
|
|
1DHG
 
 | | HG-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | MERCURY (II) ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1CFW
 
 | | GA-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | GALLIUM (III) ION, PROTEIN (DESULFOREDOXIN), SULFATE ION | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1DXG
 
 | |
2BM3
 
 | | Structure of the Type II cohesin from Clostridium thermocellum SdbA | | Descriptor: | ISOPROPYL ALCOHOL, SCAFFOLDING DOCKERIN BINDING PROTEIN A | | Authors: | Carvalho, A.L, Gloster, T.M, Pires, V.M.R, Proctor, M.R, Prates, J.A.M, Ferreira, L.M.A, Turkenburg, J.P, Romao, M.J, Davies, G.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Structural Determinants of Cohesin-Dockerin Specificity Revealed by the Crystal Structure of the Type II Cohesin from Clostridium Thermocellum Sdba.
J.Mol.Biol., 349, 2005
|
|
2C24
 
 | | FAMILY 30 CARBOHYDRATE-BINDING MODULE OF CELLULOSOMAL CELLULASE CEL9D- CEL44B OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | ENDOGLUCANASE | | Authors: | Carvalho, A.L, Alves, V.D, Najmudin, S, Romao, M.J, Prates, J.A.M, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
7Q8E
 
 | |
2CCL
 
 | | THE S45A, T46A MUTANT OF THE TYPE I COHESIN-DOCKERIN COMPLEX FROM THE CELLULOSOME OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2006-01-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evidence for a Dual Binding Mode of Dockerin Modules to Cohesins.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1OHZ
 
 | | Cohesin-Dockerin complex from the cellulosome of Clostridium thermocellum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellulosome Assembly Revealed by the Crystal Structure of the Cohesin-Dockerin Complex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OAH
 
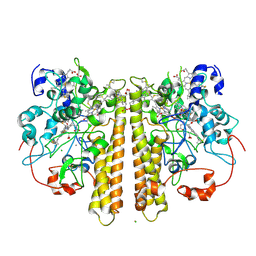 | | Cytochrome c Nitrite Reductase from Desulfovibrio desulfuricans ATCC 27774: The relevance of the two calcium sites in the structure of the catalytic subunit (NrfA). | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Cunha, C.A, Macieira, S, Dias, J.M, Almeida, G, Goncalves, L.L, Costa, C, Lampreia, J, Huber, R, Moura, J.J.G, Moura, I, Romao, M.J. | | Deposit date: | 2003-01-14 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cytochrome C Nitrite Reductase from Desulfovibrio Desulfuricans Atcc 27774. The Relevance of the Two Calcium Sites in the Structure of the Catalytic Subunit (Nrfa)
J.Biol.Chem., 278, 2003
|
|
5EPG
 
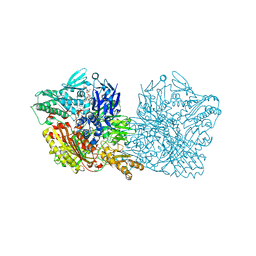 | | Human aldehyde oxidase SNP S1271L | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Coelho, C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2015-11-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Optimization of the Expression of Human Aldehyde Oxidase for Investigations of Single-Nucleotide Polymorphisms.
Drug Metab.Dispos., 44, 2016
|
|
5G5G
 
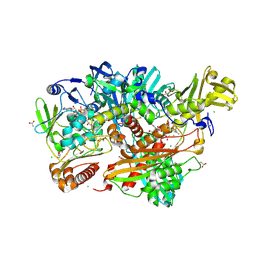 | | Escherichia coli Periplasmic Aldehyde Oxidase | | Descriptor: | ACETATE ION, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Correia, M.A.S, Otrelo-Cardoso, A.R, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Acs Chem.Biol., 11, 2016
|
|
4AO1
 
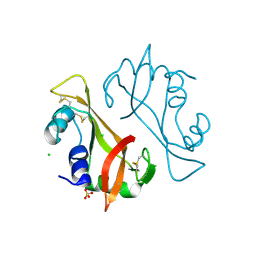 | | High resolution crystal structure of bovine pancreatic ribonuclease crystallized using ionic liquid | | Descriptor: | CHLORIDE ION, RIBONUCLEASE PANCREATIC, SULFATE ION | | Authors: | Mukhopadhyay, A, Carvalho, A.L, Kowacz, M, Romao, M.J. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Hofmeister Effects of Ionic Liquids in Protein Crystallization: Direct and Water-Mediated Interactions
Cryst.Eng.Comm., 14, 2012
|
|
4AGA
 
 | | Hofmeister effects of ionic liquids in protein crystallization: direct and water-mediated interactions | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Mukhopadhyay, A, Carvalho, A.L, Romao, M.J. | | Deposit date: | 2012-01-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hofmeister Effects of Ionic Liquids in Protein Crystallization: Direct and Water-Mediated Interactions
Cryst.Eng.Comm., 14, 2012
|
|
4AEO
 
 | | Structure of Xenobiotic Reductase B from Pseudomonas putida in complex with TNT | | Descriptor: | 2,4,6-TRINITROTOLUENE, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Carvalho, A.L, Mukhopadhyay, A, Romao, M.J, Bursakov, S, Kladova, A, Ramos, J.L, van Dillewijn, P. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of Aromatic Ring Reduction in the Xenobiotic Reductase B from Pseudomonas Putida
To be Published
|
|
5G5H
 
 | | Escherichia coli Periplasmic Aldehyde Oxidase R440H mutant | | Descriptor: | ACETATE ION, Aldehyde oxidoreductase FAD-binding subunit PaoB, Aldehyde oxidoreductase iron-sulfur-binding subunit PaoA, ... | | Authors: | Correia, M.A.S, Otrelo-Cardoso, A.R, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Acs Chem.Biol., 11, 2016
|
|
4AYZ
 
 | | X-ray Structure of human SOUL | | Descriptor: | HEME-BINDING PROTEIN 2 | | Authors: | Freire, F, Carvalho, A.L, Aveiro, S.S, Charbonnier, P, Moulis, J.M, Romao, M.J, Goodfellow, B.J, Macedo, A.L. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Human Soul: A Heme-Binding or a Bh3 Domain-Containing Protein
To be Published
|
|
4B0Y
 
 | | Determination of X-ray Structure of human SOUL by Molecular Replacement | | Descriptor: | HEME-BINDING PROTEIN 2 | | Authors: | Freire, F, Carvalho, A.L, Aveiro, S.S, Charbonnier, P, Moulis, J.M, Romao, M.J, Goodfellow, B.J, Macedo, A.L. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Human Soul: A Heme-Binding or a Bh3 Domain-Containing Protein
To be Published
|
|
4AB4
 
 | | Structure of Xenobiotic Reductase B from Pseudomonas putida in complex with TNT | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-TRINITROTOLUENE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Carvalho, A.L, Mukhopadhyay, A, Romao, M.J, Bursakov, S, Kladova, A, Ramos, J.L, van Dillewijn, P. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants of Aromatic Ring Reduction in the Xenobiotic Reductase B from Pseudomonas Putida
To be Published
|
|
2IV2
 
 | | Reinterpretation of reduced form of formate dehydrogenase H from E. coli | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase H, GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, ... | | Authors: | Raaijmakers, H.C.A, Romao, M.J. | | Deposit date: | 2006-06-08 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Formate-Reduced E. Coli Formate Dehydrogenase H: The Reinterpretation of the Crystal Structure Suggests a New Reaction Mechanism.
J.Biol.Inorg.Chem., 11, 2006
|
|
4C80
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with hydrogen peroxide | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic and Structural Studies of Aldehyde Oxidoreductase from Desulfovibrio Gigas Reveal a Dithiolene-Based Chemistry for Enzyme Activation and Inhibition by H2O2.
Plos One, 8, 2013
|
|
4BWU
 
 | | Three-dimensional structure of the K109A mutant of Paracoccus pantotrophus pseudoazurin at pH 5.5 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
