7TCX
 
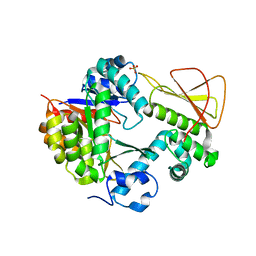 | | Methanobactin biosynthetic protein complex of MbnB and MbnC from Methylosinus trichosporium OB3b at 2.21 Angstrom resolution | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Park, Y, Reyes, R.M, Rosenzweig, A.C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TCU
 
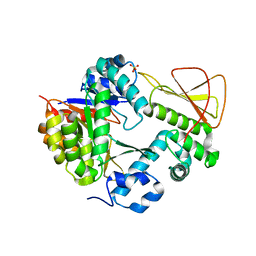 | | Methanobactin biosynthetic protein complex of MbnB and MbnC from Methylosinus trichosporium OB3b at 2.31 Angstrom resolution | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Park, Y, Reyes, R.M, Rosenzweig, A.C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4DNI
 
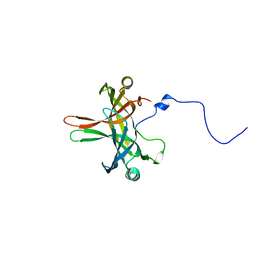 | | Structure of Editosome protein | | Descriptor: | Fusion protein of RNA-editing complex proteins MP42 and MP18 | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds.
J.Struct.Biol., 180, 2012
|
|
4DK3
 
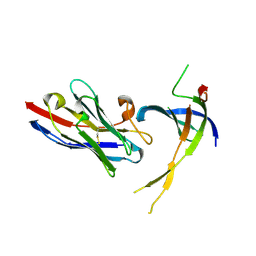 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DK6
 
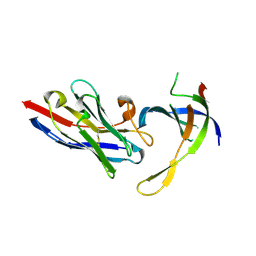 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
3STB
 
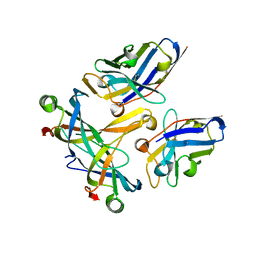 | | A complex of two editosome proteins and two nanobodies | | Descriptor: | MP18 RNA editing complex protein, RNA-editing complex protein MP42, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody.
Nucleic Acids Res., 40, 2012
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XI6
 
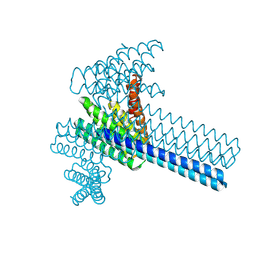 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | Descriptor: | helical fusion design | | Authors: | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XH5
 
 | |
6XT4
 
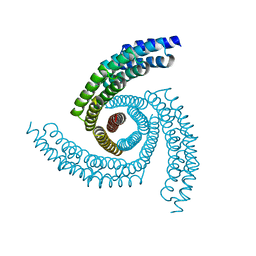 | |
6XNS
 
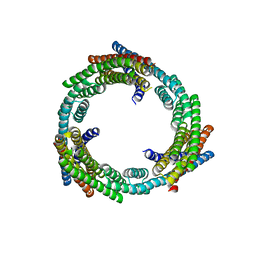 | | C3_crown-05 | | Descriptor: | C3_crown-05 | | Authors: | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6VFH
 
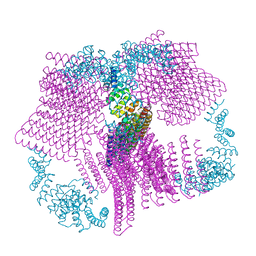 | |
6VFJ
 
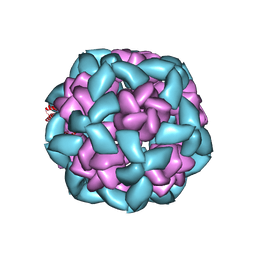 | |
6VFI
 
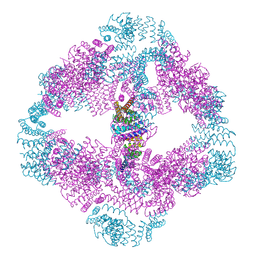 | |
6V8E
 
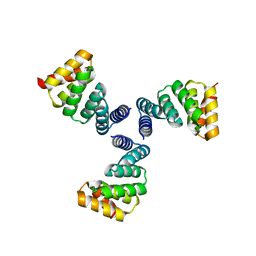 | |
6VEH
 
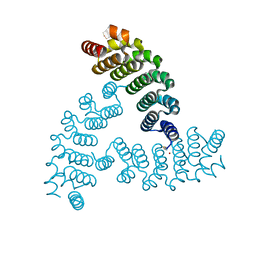 | |
8TVI
 
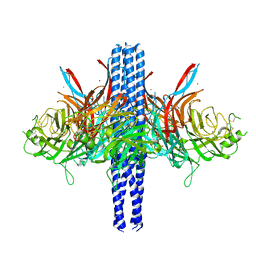 | |
8TVG
 
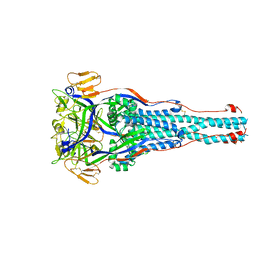 | |
8TVF
 
 | |
8TVE
 
 | |
8TVH
 
 | | Langya henipavirus postfusion F protein in complex with 4G5 Fab, local refinement of the viral membrane proximal region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4G5 heavy chain, 4G5 light chain, ... | | Authors: | Wang, Z, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and design of Langya virus glycoprotein antigens.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
