7GTK
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000552a | | Descriptor: | (4R)-2-(2-hydroxyethyl)-4-methoxy-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTP
 
 | |
7GTU
 
 | |
7GTX
 
 | |
7GTW
 
 | |
7GU0
 
 | |
7GS9
 
 | |
7GSE
 
 | |
7GSG
 
 | |
7GU2
 
 | |
7GU3
 
 | |
7GTF
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000754b | | Descriptor: | 1,2-benzoxazol-3-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSK
 
 | |
7GSQ
 
 | |
7GTO
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000602a | | Descriptor: | (6aR,8R,12R,12aS)-5-methyl-5,6a,7,10,11,12a-hexahydro-6H,9H-pyrazolo[1',2':1,2]pyrazolo[4,3-c]quinolin-9-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTD
 
 | |
7GSU
 
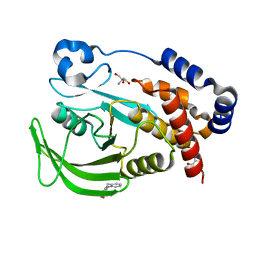 | |
7GTR
 
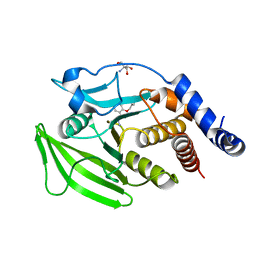 | |
7GSY
 
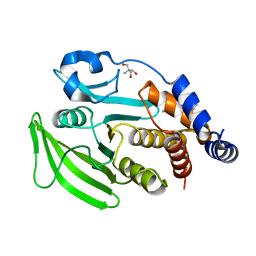 | |
7GT5
 
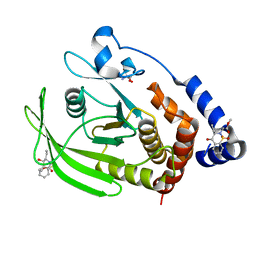 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000529a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, methyl [(3R,4S)-3-ethyl-4-hydroxy-1,1-dioxo-3,4-dihydro-1lambda~6~,2-benzothiazin-2(1H)-yl]acetate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTS
 
 | |
7GSR
 
 | |
7GS7
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000621a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1,2,3-thiadiazol-4-yl)phenyl ethylcarbamate, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTT
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000148a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(3,4-dihydroquinoline-1(2H)-carbothioyl)propanamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSA
 
 | |
