3HJI
 
 | |
5F68
 
 | |
6P41
 
 | |
6P43
 
 | |
6P42
 
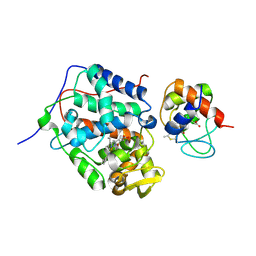 | |
1U0S
 
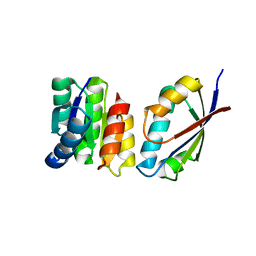 | | Chemotaxis kinase CheA P2 domain in complex with response regulator CheY from the thermophile thermotoga maritima | | Descriptor: | Chemotaxis protein cheA, Chemotaxis protein cheY | | Authors: | Park, S.Y, Beel, B.D, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TQG
 
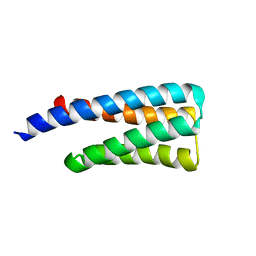 | | CheA phosphotransferase domain from Thermotoga maritima | | Descriptor: | Chemotaxis protein cheA | | Authors: | Quezada, C.M, Gradinaru, C, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima.
J.Mol.Biol., 341, 2004
|
|
8DIK
 
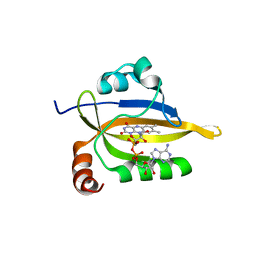 | |
5TDY
 
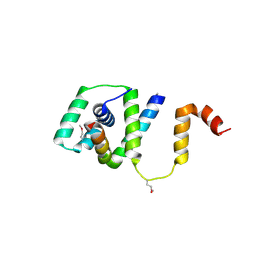 | | Structure of cofolded FliFc:FliGn complex from Thermotoga maritima | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG | | Authors: | Lynch, M.J, Levenson, R, Kim, E.A, Sircar, R, Blair, D.F, Dahlquist, F.W, Crane, B.R. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Co-Folding of a FliF-FliG Split Domain Forms the Basis of the MS:C Ring Interface within the Bacterial Flagellar Motor.
Structure, 25, 2017
|
|
2I7O
 
 | | Structure of Re(4,7-dimethyl-phen)(Thr124His)(Lys122Trp)(His83Gln)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Sudhamsu, J, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tryptophan-accelerated electron flow through proteins.
Science, 320, 2008
|
|
2I7S
 
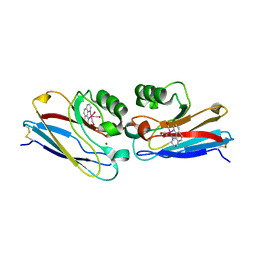 | | Crystal structure of Re(phen)(CO)3 (Thr124His)(His83Gln) Azurin Cu(II) from Pseudomonas aeruginosa | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COBALT TETRAAMMINE ION, ... | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
3RH8
 
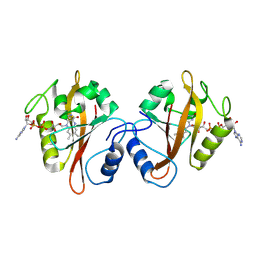 | |
3T8Y
 
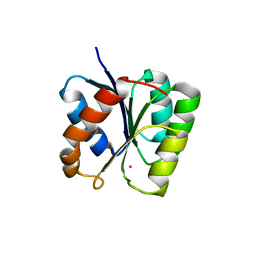 | |
3RTY
 
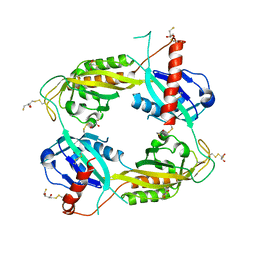 | | Structure of an Enclosed Dimer Formed by The Drosophila Period Protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Period circadian protein | | Authors: | King, H.A, Hoelz, A, Crane, B.R, Young, M.W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of an enclosed dimer formed by the Drosophila period protein.
J.Mol.Biol., 413, 2011
|
|
3SFT
 
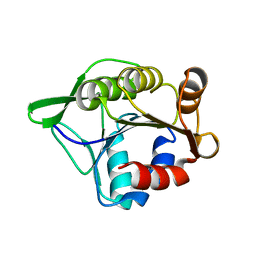 | |
3SOH
 
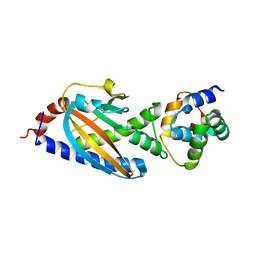 | | Architecture of the Flagellar Rotor | | Descriptor: | Flagellar motor switch protein FliG, Flagellar motor switch protein FliM | | Authors: | Koushik, P, Gonzalez-Bonet, G, Bilwes, A.M, Crane, B.R, Blair, D. | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the flagellar rotor.
Embo J., 30, 2011
|
|
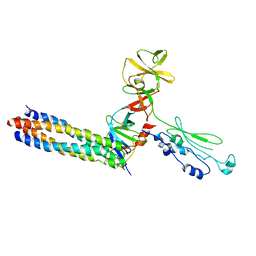
3UR1
 
 | |
4HI4
 
 | |
4I44
 
 | |
4I3M
 
 | | Aer2 poly-HAMP domains: L44H HAMP1 CW-lock mutant | | Descriptor: | Aerotaxis transducer Aer2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Airola, M.V, Sukomon, N, Crane, B.R. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HAMP Domain Conformers That Propagate Opposite Signals in Bacterial Chemoreceptors.
Plos Biol., 11, 2013
|
|
4HZ1
 
 | | Crystal Structure of Pseudomonas aeruginosa azurin with iron(II) at the copper-binding site. | | Descriptor: | ACETATE ION, Azurin, FE (II) ION | | Authors: | McLaughlin, M.P, Retegan, M, Bill, E, Payne, T.M, Shafaat, H.S, Pea, S, Sudhamsu, J, Ensign, A.A, Crane, B.R, Neese, F, Holland, P.L. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azurin as a Protein Scaffold for a Low-coordinate Nonheme Iron Site with a Small-molecule Binding Pocket.
J.Am.Chem.Soc., 134, 2012
|
|
4JPB
 
 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
1R1C
 
 | | PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(HIS107) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (I) ION | | Authors: | Miller, J.E, Gradinaru, C, Crane, B.R, Di Bilio, A.J. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spectroscopy and reactivity of a photogenerated tryptophan radical in a structurally defined protein environment
J.Am.Chem.Soc., 125, 2003
|
|
1XKR
 
 | | X-ray Structure of Thermotoga maritima CheC | | Descriptor: | chemotaxis protein CheC | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX
Mol.Cell, 16, 2004
|
|
1XKO
 
 | | Structure of Thermotoga maritima CheX | | Descriptor: | CHEMOTAXIS protein cheX | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX.
Mol.Cell, 16, 2004
|
|
