5LTQ
 
 | |
5LNT
 
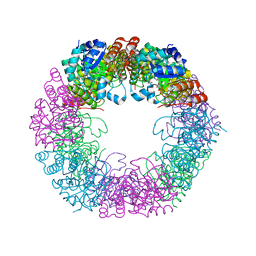 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LTR
 
 | |
2YM1
 
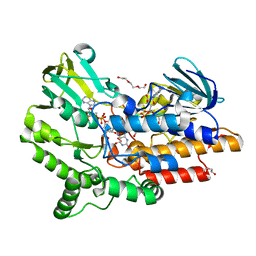 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Arg337Lys MUTANT IN COMPLEX WITH NADP | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YLS
 
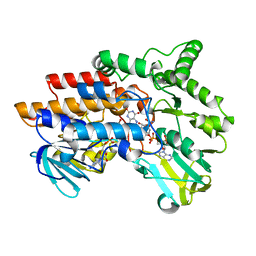 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: REDUCED ENZYME BOUND TO NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YM2
 
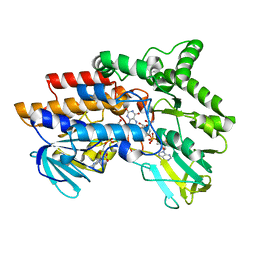 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Arg337Lys MUTANT REDUCED STATE WITH NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
6QQK
 
 | |
6QQI
 
 | |
6QQJ
 
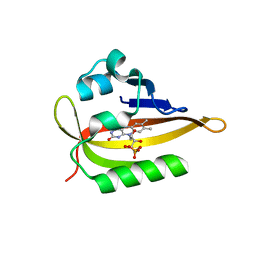 | |
6QSA
 
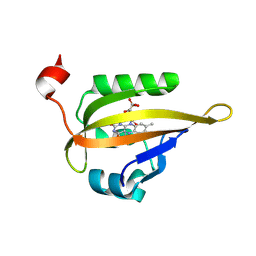 | |
2YLZ
 
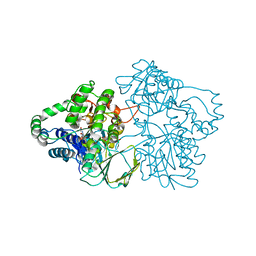 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Met446Gly MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHENYLACETONE MONOOXYGENASE, SULFATE ION | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YLW
 
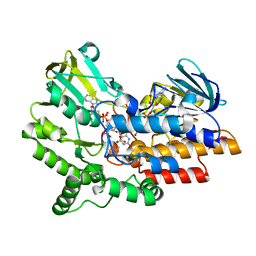 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Arg337Lys MUTANT IN COMPLEX WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YLR
 
 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: COMPLEX WITH NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YLT
 
 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: COMPLEX WITH NADP and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YLX
 
 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Asp66Ala MUTANT IN COMPLEX WITH NADP AND MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7Z7P
 
 | |
7Z7O
 
 | |
7Z7Q
 
 | |
6S68
 
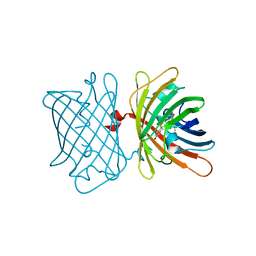 | | Structure of the Fluorescent Protein AausFP2 from Aequorea cf. australis at pH 7.6 | | Descriptor: | Aequorea cf. australis fluorescent protein 2 (AausFP2) | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
6S67
 
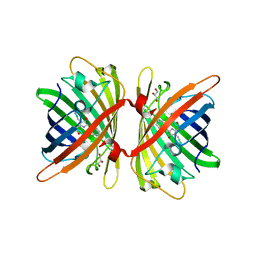 | | Structure of the Fluorescent Protein AausFP1 from Aequorea cf. australis at pH 7.0 | | Descriptor: | Aequorea cf. australis fluorescent protein 1 (AausFP1), GLYCEROL | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
