7VHP
 
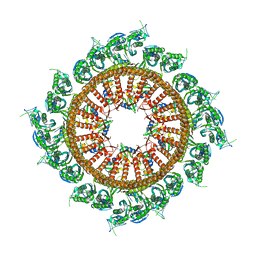 | | Structural insights into the membrane microdomain organization by SPFH family proteins | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Protein HflK | | Authors: | Ma, C.Y, Wang, C.K, Luo, D.Y, Yan, L, Yang, W.X, Li, N.N, Gao, N. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the membrane microdomain organization by SPFH family proteins.
Cell Res., 32, 2022
|
|
8HDV
 
 | |
8HDU
 
 | |
7YD9
 
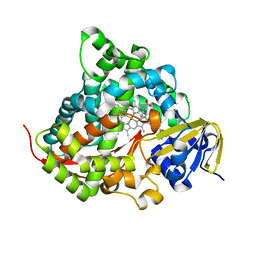 | | Crystal structure of the P450 BM3 heme domain mutant F87G/T268V/A184V/A328V in complex with N-imidazolyl-hexanoyl-L-phenylalanine,methylbenzene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDA
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87V/T268V/A184V in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDB
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87V/T268I in complex with N-imidazolyl-pentanoyl-L-phenylalanine,ethylbenzene and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268P/V78I in complex with N-imidazolyl-pentanoyl-L-phenylalanine,propylbenzene and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, HYDROXYAMINE, P450 BM3 heme domain mutant F87A/T268P/V78I, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDC
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L/T268V/V78C in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJF
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268P/V78I in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, HYDROXYAMINE, P450 BM3 heme domain mutant F87A/T268P/V78I, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDE
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87T/T268V/I263V in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YFT
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine, indane and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, 2,3-dihydro-1H-indene, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJE
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87G/T268V/A184V/A328V in complex with N-imidazolyl-hexanoyl-L-phenylalanine and acetate ion | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, ACETATE ION, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YJG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
9D53
 
 | | PAK4 in complex with compound 7 | | Descriptor: | N~2~-{[(1s,4s)-4-aminocyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 4 | | Authors: | Boone, C.D, Olland, A.M, Suto, R.K. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
8FA1
 
 | |
8FA2
 
 | |
4DRX
 
 | | GTP-Tubulin in complex with a DARPIN | | Descriptor: | Designed ankyrin repeat protein (DARPIN) D1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pecqueur, L, Gigant, B, Knossow, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KB5
 
 | | Crystal structure of MycP1 from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Tian, C.L. | | Deposit date: | 2013-04-23 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
9JUY
 
 | | X-ray crystal structure of Y16513 in CBP | | Descriptor: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9JUT
 
 | | X-ray crystal structure of Y16524 in EP300 | | Descriptor: | (6~{S})-1-(3-chloranyl-4-methoxy-phenyl)-6-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]piperidin-2-one, Histone acetyltransferase p300 | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9JUU
 
 | | X-ray crystal structure of Y16515 in CBP | | Descriptor: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{R})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
7WMW
 
 | |
7WMY
 
 | |
7WMX
 
 | |
7WMZ
 
 | |
