5KH9
 
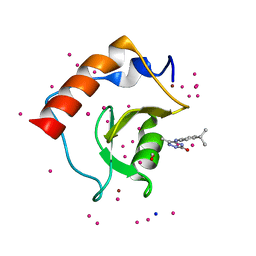 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
6YK7
 
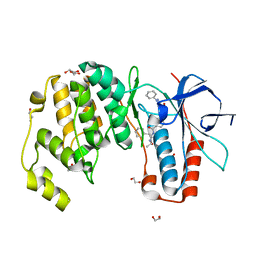 | | Crystal structure of p38 in complex with SR43 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(ethylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y4W
 
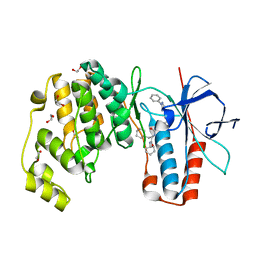 | | Crystal structure of p38 in complex with SR69 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(cyclohexylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6YAT
 
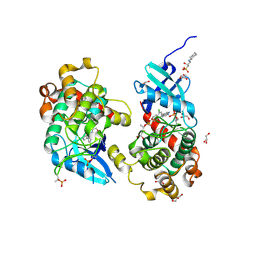 | | Crystal structure of STK4 (MST1) in complex with compound 6 | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 4-[5-(3-chlorophenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]morpholine, GLYCEROL, ... | | Authors: | Chaikuad, A, Bata, N, Limpert, A.S, Lambert, L.J, Bakas, N.A, Cosford, N.D.P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Inhibitors of the Hippo Pathway Kinases STK3/MST2 and STK4/MST1 Have Utility for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 65, 2022
|
|
5ENF
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with fragment-4 N10142 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-2-(2-methylpropyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
6Y4X
 
 | | Crystal structure of p38 in complex with SR72 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[2-(4-chlorophenyl)ethylamino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6YTD
 
 | | CLK1 V324A mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTI
 
 | | CLK1 bound with ETH1610 (Cpd 17) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
5ENH
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-12 N11528 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[(2,6-dimethoxyphenyl)methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
6YU1
 
 | | CLK3 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTA
 
 | | CLK1 bound with imidazopyridazine (Cpd 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTG
 
 | | CLK1 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Huber, K, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
5IFU
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with Glyburide | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, CHLORIDE ION, ... | | Authors: | Dranow, D.M, Hewitt, S.N, Abendroth, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Selective Allosteric Inhibitors of the Plasmodium falciparum Drug Target, Prolyl-tRNA-synthetase.
ACS Infect Dis, 3, 2017
|
|
6ZED
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dimethyl-1~{H}-pyrazolo[3,4-c]pyridazin-3-amine, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1
To Be Published
|
|
6YTW
 
 | | CLK3 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTY
 
 | | CLK3 A319V mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
5ENB
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with o-Tolylthiourea (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-methylphenyl)thiourea, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENI
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-13 N11537 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-2-oxidanyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5LVR
 
 | | Crystal structure of human PCAF bromodomain in complex with compound-E (CPD-E) | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-phenyl-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | Descriptor: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | Authors: | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-23 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
5ENE
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with 5-Amino-2-benzyl-1,3-oxazole-4-carbonitrile (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 5-azanyl-2-(phenylmethyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENJ
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-14 N11530 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | MAGNESIUM ION, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-~{N}-methyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
6ZF9
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4
To Be Published
|
|
6YTE
 
 | | CLK1 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
