4DIB
 
 | | The crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Bacillus anthracis str. Sterne | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | Authors: | Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Bacillus anthracis str. Sterne
To be Published
|
|
4HDE
 
 | | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames | | Descriptor: | SCO1/SenC family lipoprotein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames
To be Published
|
|
4EBJ
 
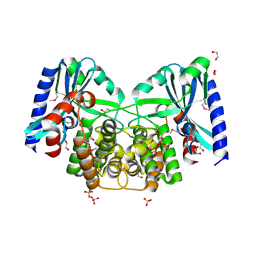 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo
To be Published
|
|
4EG9
 
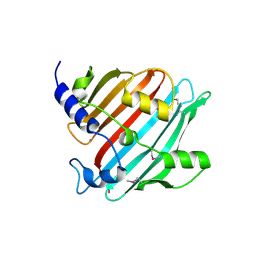 | | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
4EH1
 
 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
4E4R
 
 | | EutD phosphotransacetylase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | EutD phosphotransacetylase from Staphylococcus aureus.
To be Published
|
|
4H59
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane
To be Published
|
|
4HCF
 
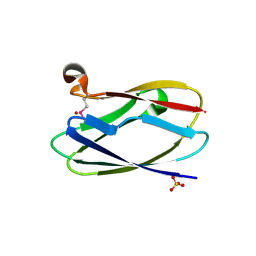 | | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Copper Bound from Bacillus anthracis | | Descriptor: | COPPER (II) ION, Cupredoxin 1, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Copper Bound from Bacillus anthracis
To be Published
|
|
4H3U
 
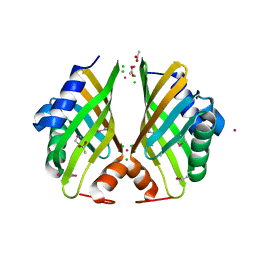 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
4HAO
 
 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92 | | Descriptor: | ACETIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92
To be Published
|
|
3O6C
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
3NXK
 
 | | Crystal Structure of Probable Cytoplasmic L-asparaginase from Campylobacter jejuni | | Descriptor: | ACETIC ACID, Cytoplasmic L-asparaginase, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Probable Cytoplasmic L-asparaginase from Campylobacter jejuni
To be Published
|
|
3NTX
 
 | | Crystal Structure of L-asparaginase I from Yersinia pestis | | Descriptor: | Cytoplasmic L-asparaginase I, GLYCEROL | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-05 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L-asparaginase I from Yersinia pestis
To be Published, 2010
|
|
3O7M
 
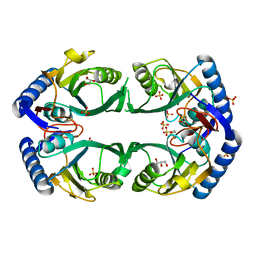 | | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor'
TO BE PUBLISHED
|
|
3NZ2
 
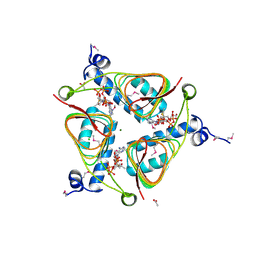 | | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, ACETYL COENZYME *A, ... | | Authors: | Kim, Y, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor
To be Published
|
|
3OT5
 
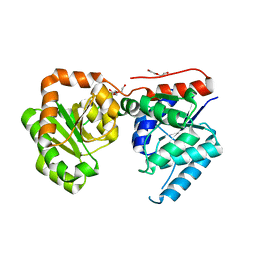 | | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes.
TO BE PUBLISHED
|
|
3PAJ
 
 | | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | MAGNESIUM ION, Nicotinate-nucleotide pyrophosphorylase, carboxylating | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3OZH
 
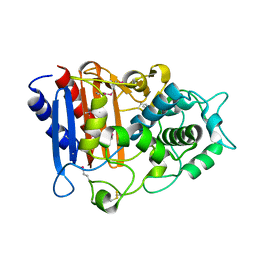 | | Crystal Structure of Beta-Lactamase/D-alanine Carboxypeptidase from Yersinia pestis | | Descriptor: | beta-lactamase/D-alanine carboxypeptidase | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-24 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-alanine Carboxypeptidase from Yersinia pestis
To be Published
|
|
3P0R
 
 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
3P2L
 
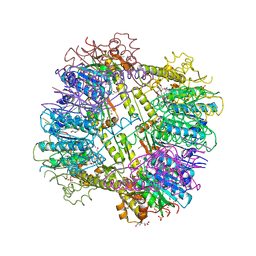 | | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-02 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis
To be Published
|
|
3OPQ
 
 | | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure. | | Descriptor: | CHLORIDE ION, FORMIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.
To be Published
|
|
3OUZ
 
 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
3QFK
 
 | | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Krishna, S.N, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate.
TO BE PUBLISHED
|
|
3QTT
 
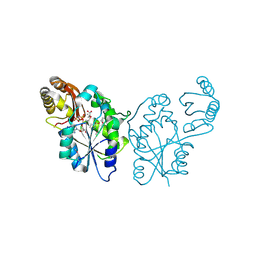 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine | | Descriptor: | BETA-ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine.
To be Published
|
|
3Q94
 
 | | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-07 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
