4LYQ
 
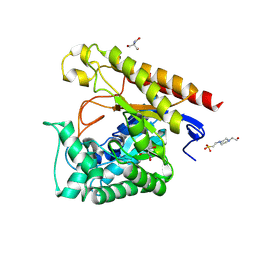 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8YFJ
 
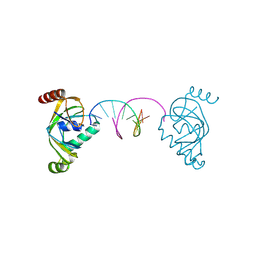 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-ATTGGATCCAAT-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*AP*TP*TP*GP*GP*AP*TP*CP*CP*AP*AP*T)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YEY
 
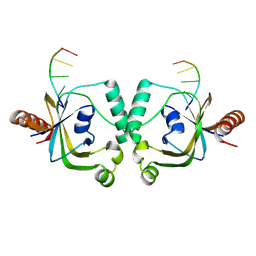 | | TRIP4 ASCH domain in complex with ssDNA-1 | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*TP*TP*TP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YFI
 
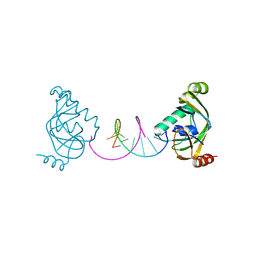 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCA-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YEW
 
 | | TRIP4 ASCH domain in unliganded form | | Descriptor: | Activating signal cointegrator 1 | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXX
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCG-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*CP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*G)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXW
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCC-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
7JVM
 
 | |
7JVN
 
 | |
1PGL
 
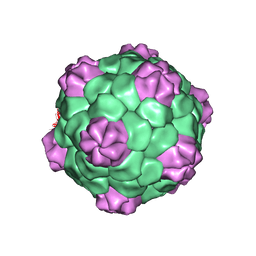 | | BEAN POD MOTTLE VIRUS (BPMV), MIDDLE COMPONENT | | Descriptor: | 5'-R(*AP*GP*UP*CP*UP*C)-3', BEAN POD MOTTLE VIRUS LARGE (L) SUBUNIT, BEAN POD MOTTLE VIRUS SMALL (S) SUBUNIT | | Authors: | Lin, T, Cavarelli, J, Johnson, J.E. | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus.
Virology, 314, 2003
|
|
1PGW
 
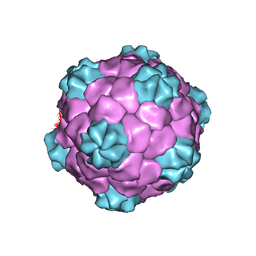 | | BEAN POD MOTTLE VIRUS (BPMV), TOP COMPONENT | | Descriptor: | BEAN POD MOTTLE VIRUS LARGE (L) SUBUNIT, BEAN POD MOTTLE VIRUS SMALL (S) SUBUNIT | | Authors: | Lin, T, Cavarelli, J, Johnson, J.E. | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus.
Virology, 314, 2003
|
|
8J5J
 
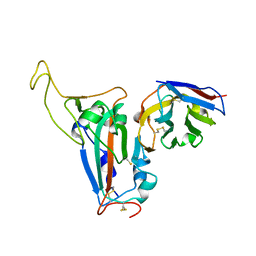 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
6M6U
 
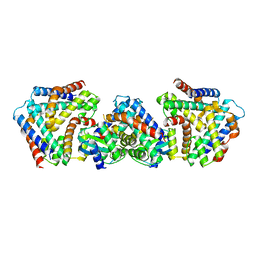 | | Crystal structure the toxin-antitoxin MntA-HpeT mutant-D39ED41E | | Descriptor: | Toxin-antitoxin system antitoxin MntA family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5I89
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02857790 | | Descriptor: | (4R)-6-(3-cyclopropyl-1-methyl-1H-indazol-5-yl)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Setser, J.W, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I83
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02773986 | | Descriptor: | (4R)-4-methyl-7-[(1R)-1-phenylethoxy]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein, THIOCYANATE ION | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5EHR
 
 | |
5EHP
 
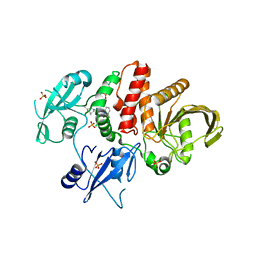 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP836 | | Descriptor: | 5-[2,3-bis(chloranyl)phenyl]-2-[(3~{R},5~{S})-3,5-dimethylpiperazin-1-yl]pyrimidin-4-amine, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5I8B
 
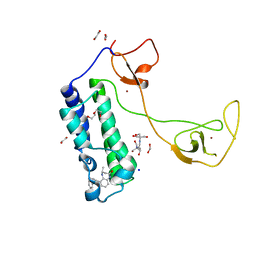 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I8G
 
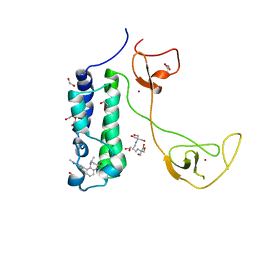 | | CBP in complex with Cpd637 ((R)-4-methyl-6-(1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-4-methyl-6-[1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I86
 
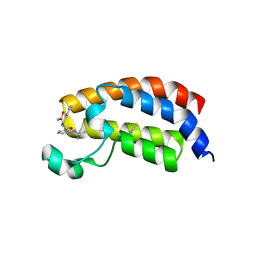 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
