5KAO
 
 | | Crystal structure of wild type HIV-1 protease in complex with GRL-10413 | | Descriptor: | [(3~{a}~{S},4~{R},6~{a}~{R})-2,3,3~{a},4,5,6~{a}-hexahydrofuro[2,3-b]furan-4-yl] ~{N}-[(2~{S},3~{R})-1-(3-chloranyl-4-methoxy-phenyl)-4-[(4-methoxyphenyl)sulfonyl-(2-methylpropyl)amino]-3-oxidanyl-butan-2-yl]carbamate, protease | | Authors: | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-06-01 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.
Antimicrob.Agents Chemother., 60, 2016
|
|
3GB1
 
 | | STRUCTURES OF B1 DOMAIN OF STREPTOCOCCAL PROTEIN G | | Descriptor: | PROTEIN (B1 DOMAIN OF STREPTOCOCCAL PROTEIN G) | | Authors: | Clore, G.M. | | Deposit date: | 1999-05-02 | | Release date: | 1999-06-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Improving the Packing and Accuracy of NMR Structures with a Pseudopotential for the Radius of Gyration
J.Am.Chem.Soc., 121, 1999
|
|
1GB1
 
 | |
2GB1
 
 | |
7S21
 
 | | M. xanthus encapsulin shell protein EncA with T=1 symmetry | | Descriptor: | EncA | | Authors: | Eren, E. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
7S4Q
 
 | | M. xanthus encapsulin EncA bound to EncC targeting peptide | | Descriptor: | EncA, EncC targeting peptide | | Authors: | Eren, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
7S20
 
 | | M. xanthus encapsulin shell protein EncA with T=3 symmetry | | Descriptor: | EncA | | Authors: | Eren, E. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
7S8T
 
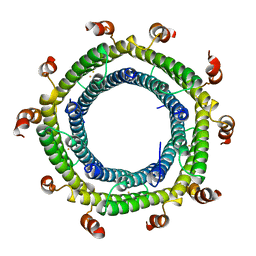 | | M. xanthus ferritin-like protein EncC | | Descriptor: | EncC, FE (III) ION | | Authors: | Eren, E. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
7S5C
 
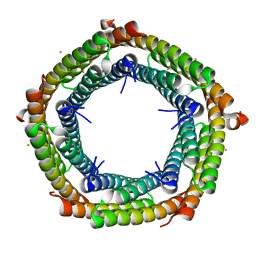 | | M. xanthus ferritin-like protein EncB | | Descriptor: | CALCIUM ION, EncB, FE (III) ION | | Authors: | Eren, E. | | Deposit date: | 2021-09-10 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
7S5K
 
 | | M. xanthus ferritin-like protein EncB | | Descriptor: | CALCIUM ION, EncB, FE (III) ION | | Authors: | Eren, E. | | Deposit date: | 2021-09-10 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
7S2T
 
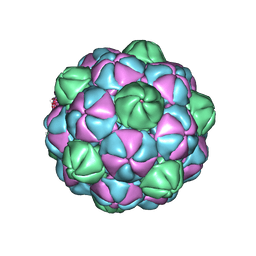 | | M. xanthus encapsulin EncA bound to EncB targeting peptide | | Descriptor: | EncA, EncB targeting peptide | | Authors: | Eren, E. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism.
Structure, 30, 2022
|
|
1MPE
 
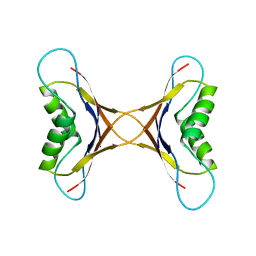 | |
1NER
 
 | |
1MVK
 
 | | X-ray structure of the tetrameric mutant of the B1 domain of streptococcal protein G | | Descriptor: | Immunoglobulin G binding protein G, SULFATE ION | | Authors: | Frank, M.K, Dyda, F, Dobrodumov, A, Gronenborn, A.M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Core mutations switch monomeric protein GB1 into an intertwined tetramer.
Nat.Struct.Biol., 9, 2002
|
|
1NEQ
 
 | |
1D8V
 
 | | THE RESTRAINED AND MINIMIZED AVERAGE NMR STRUCTURE OF MAP30. | | Descriptor: | ANTI-HIV AND ANTI-TUMOR PROTEIN MAP30 | | Authors: | Wang, Y.-X, Neamati, N, Jacob, J, Palmer, I, Stahl, S.J. | | Deposit date: | 1999-10-26 | | Release date: | 1999-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of anti-HIV-1 and anti-tumor protein MAP30: structural insights into its multiple functions.
Cell(Cambridge,Mass.), 99, 1999
|
|
1IGD
 
 | |
1IGC
 
 | |
1PGB
 
 | |
1PGA
 
 | |
1Q10
 
 | |
1PGX
 
 | |
1CQH
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
1CQG
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, 31 STRUCTURES | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
1AVV
 
 | | HIV-1 NEF PROTEIN, UNLIGANDED CORE DOMAIN | | Descriptor: | NEGATIVE FACTOR | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1997-09-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling.
Structure, 5, 1997
|
|
