3TLT
 
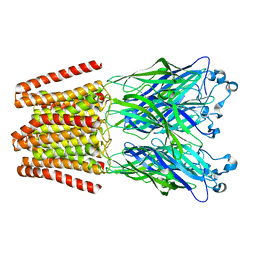 | | The GLIC pentameric Ligand-Gated Ion Channel H11'F mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4IL9
 
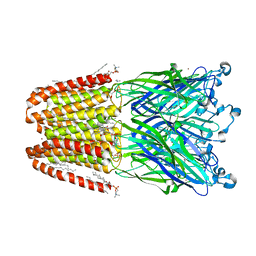 | | The pentameric ligand-gated ion channel GLIC A237F in complex with bromide | | Descriptor: | ACETATE ION, BROMIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILB
 
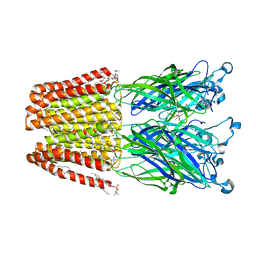 | | The pentameric ligand-gated ion channel GLIC A237F in complex with Rubidium | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILC
 
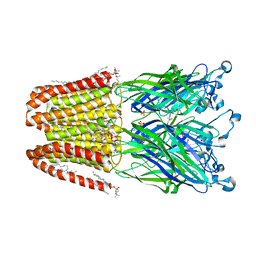 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4HFI
 
 | | The GLIC pentameric Ligand-Gated Ion Channel at 2.4 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILA
 
 | | The pentameric ligand-gated ion channel GLIC A237F in complex with Cesium | | Descriptor: | ACETATE ION, CESIUM ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4IL4
 
 | | The pentameric ligand-gated ion channel GLIC in complex with Se-DDM | | Descriptor: | ACETATE ION, CHLORIDE ION, Proton-gated ion channel, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
8SCY
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8TSZ
 
 | |
8TSY
 
 | |
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT0
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSU
 
 | |
8TSX
 
 | |
8TT1
 
 | |
7VJZ
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 1.90 A resolution-7 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Tokay, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK7
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.4 A resolution-11 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Dag, C. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK0
 
 | |
7VK2
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.0 A resolution -9 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Gul, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK8
 
 | | Crystal structure of SARS-CoV-2 Mpro at 2.4 A Resolution | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Usta, G. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK1
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 1.93 A resolution-5 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Ertem, B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK6
 
 | |
7VJY
 
 | |
