7VJG
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (1 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ1
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (10 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ5
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (5 ms time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJB
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJH
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (5 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VIY
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (50 ns time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ4
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (1 ms time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJE
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (300 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJI
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (10 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ0
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (1 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJA
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (10 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VIX
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (10 ns time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJJ
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AGA
 
 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ANS
 
 | | Structure of SARS-CoV-2 Main Protease bound to Adrafinil. | | Descriptor: | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AVD
 
 | | Structure of SARS-CoV-2 Main Protease bound to SEN1269 ligand | | Descriptor: | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol, 3C-like proteinase, CHLORIDE ION | | Authors: | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWU
 
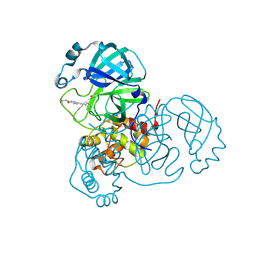 | | Structure of SARS-CoV-2 Main Protease bound to LSN2463359. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, ~{N}-propan-2-yl-5-(2-pyridin-4-ylethynyl)pyridine-2-carboxamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AF0
 
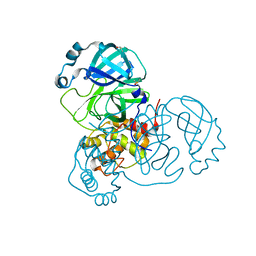 | | Structure of SARS-CoV-2 Main Protease bound to Ipidacrine. | | Descriptor: | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7APH
 
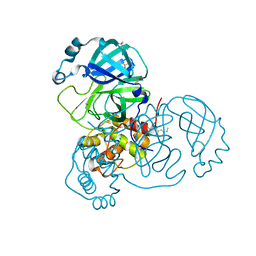 | | Structure of SARS-CoV-2 Main Protease bound to Tofogliflozin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AK4
 
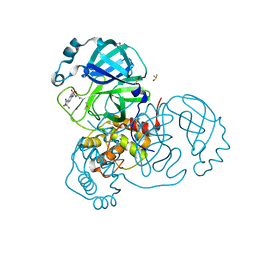 | | Structure of SARS-CoV-2 Main Protease bound to Tretazicar. | | Descriptor: | 3C-like proteinase, 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AMJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to PD 168568. | | Descriptor: | (3~{S})-3-[2-[4-(3,4-dimethylphenyl)piperazin-1-yl]ethyl]-2,3-dihydroisoindol-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AKU
 
 | | Structure of SARS-CoV-2 Main Protease bound to Calpeptin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Calpeptin | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-02 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AX6
 
 | | Structure of SARS-CoV-2 Main Protease bound to Glutathione isopropyl ester | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
