1IX3
 
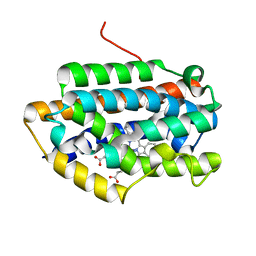 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Cyanide | | Descriptor: | CYANIDE ION, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1UBB
 
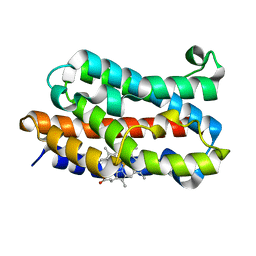 | | Crystal structure of rat HO-1 in complex with ferrous heme | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Higashimoto, Y, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1ULX
 
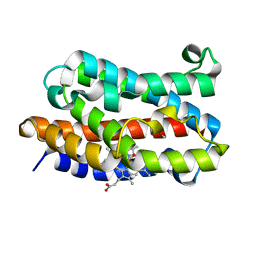 | | Partially photolyzed structure of CO-bound heme-heme oxygenase complex | | Descriptor: | CARBON MONOXIDE, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-09-16 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
1VGI
 
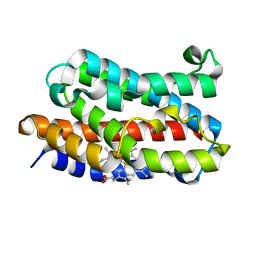 | | Crystal structure of xenon bound rat heme-heme oxygenase-1 complex | | Descriptor: | FORMIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
1HSS
 
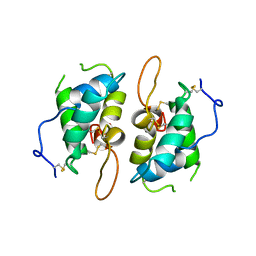 | | 0.19 ALPHA-AMYLASE INHIBITOR FROM WHEAT | | Descriptor: | 0.19 ALPHA-AMYLASE INHIBITOR | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution.
Biochemistry, 36, 1997
|
|
1IR6
 
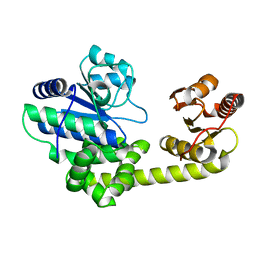 | | Crystal structure of exonuclease RecJ bound to manganese | | Descriptor: | MANGANESE (II) ION, exonuclease RecJ | | Authors: | Yamagata, A, Kakuta, Y, Masui, R, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1D2M
 
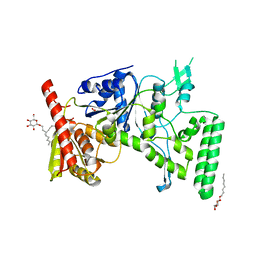 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
1DVE
 
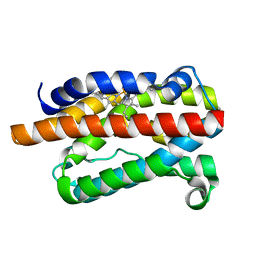 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
1ARP
 
 | |
1UJ8
 
 | |
2E0Y
 
 | |
2E0X
 
 | |
2E0W
 
 | |
1J02
 
 | | Crystal Structure of Rat Heme Oxygenase-1-Heme Bound to NO | | Descriptor: | HEME OXYGENASE 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
3AV8
 
 | | Refined Structure of Plant-type [2Fe-2S] Ferredoxin I from Aphanothece sacrum at 1.46 A Resolution | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, SULFATE ION | | Authors: | Kameda, H, Hirabayashi, K, Wada, K, Fukuyama, K. | | Deposit date: | 2011-02-28 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Mapping of protein-protein interaction sites in the plant-type [2Fe-2S] ferredoxin.
Plos One, 6, 2011
|
|
1J2C
 
 | | Crystal structure of rat heme oxygenase-1 in complex with biliverdin IXalpha-iron cluster | | Descriptor: | BILIVERDINE IX ALPHA, FE (III) ION, Heme Oxygenase-1 | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-12-29 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Rat Heme Oxygenase-1 in Complex with Biliverdin-Iron Chelate: CONFORMATIONAL CHANGE OF THE DISTAL HELIX DURING THE HEME CLEAVAGE REACTION.
J.Biol.Chem., 278, 2003
|
|
2Z8I
 
 | |
2DBW
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Acyl-Enzyme Intermediate | | Descriptor: | GAMMA-L-GLUTAMIC ACID, GLYCEROL, Gamma-glutamyltranspeptidase | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2DBX
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Complexed with L-Glutamate | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DBU
 
 | |
2E0N
 
 | |
2Z7E
 
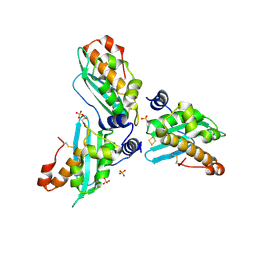 | | Crystal structure of Aquifex aeolicus IscU with bound [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, NifU-like protein, SULFATE ION | | Authors: | Shimomura, Y, Wada, K, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2007-08-20 | | Release date: | 2008-08-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis
J.Mol.Biol., 383, 2008
|
|
1I7H
 
 | | CRYSTAL STURCUTURE OF FDX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Kakuta, Y, Horio, T, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2001-03-09 | | Release date: | 2002-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli Fdx, an adrenodoxin-type ferredoxin involved in the assembly of iron-sulfur clusters.
Biochemistry, 40, 2001
|
|
2Z8J
 
 | |
