8E23
 
 | | Human DNA polymerase theta in complex with allosteric inhibitor | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*TP*CP*CP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*C)-3'), DNA (5'-D(*GP*C*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*G)-3'), ... | | Authors: | Mader, P, Pau, V.P.T, Sicheri, F. | | Deposit date: | 2022-08-13 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Identification of RP-6685 , an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Pol theta.
J.Med.Chem., 65, 2022
|
|
8E24
 
 | | Human DNA polymerase theta in complex with allosteric inhibitor | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-[2,4-bis(trifluoromethyl)phenyl]-N-phenyl-N-[3-(pyridazin-3-yl)prop-2-yn-1-yl]acetamide, DNA, ... | | Authors: | Mader, P, Pau, V.P.T, Sicheri, F. | | Deposit date: | 2022-08-13 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of RP-6685 , an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Pol theta.
J.Med.Chem., 65, 2022
|
|
8T8D
 
 | | Structure of Helicobacter pylori adhesin A, HpaA | | Descriptor: | ACETATE ION, GLYCEROL, Neuraminyllactose-binding hemagglutinin | | Authors: | Martini, C, Calmettes, C. | | Deposit date: | 2023-06-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the crystal structure of the HpaA adhesin: insights into cell adhesion function and epitope localization of a Helicobacter pylori vaccine candidate.
Mbio, 15, 2024
|
|
3N75
 
 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | Authors: | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | Deposit date: | 2010-05-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
8US2
 
 | |
8US1
 
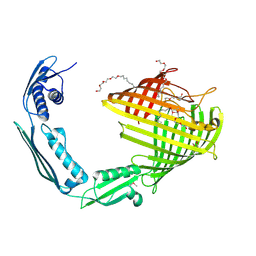 | |
8US4
 
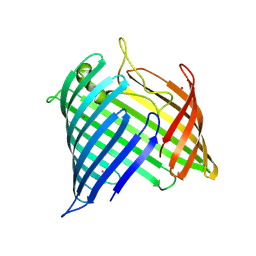 | |
8US3
 
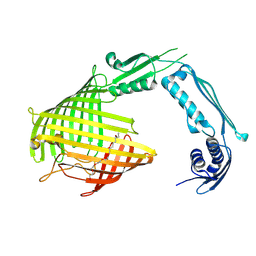 | |
4RVW
 
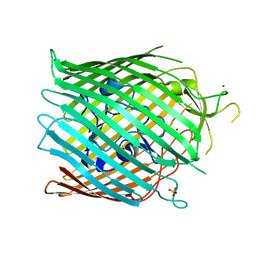 | |
4RDR
 
 | |
4JLG
 
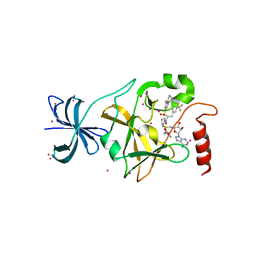 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5FL2
 
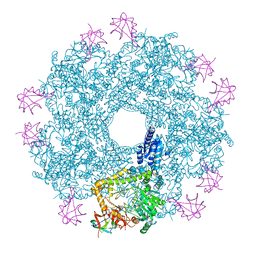 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
4HC4
 
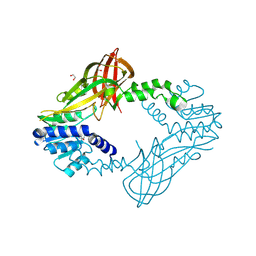 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4NW2
 
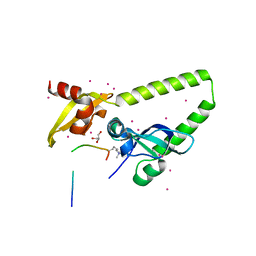 | | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Tempel, W, Xu, C, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
