2Q8K
 
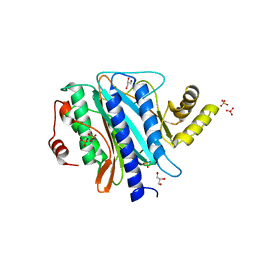 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
2QY9
 
 | |
7O0N
 
 | |
6TWJ
 
 | |
6TWM
 
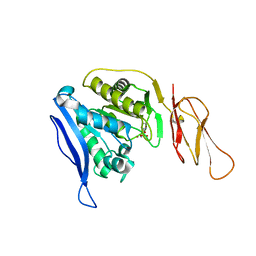 | | Product bound structure of the Ectoine utilization protein EutE (DoeB) from Ruegeria pomeroyi | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, ACETATE ION, N-acetyl-L-2,4-diaminobutyric acid deacetylase, ... | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
6TWK
 
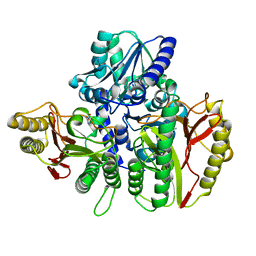 | | Substrate bound structure of the Ectoine utilization protein EutD (DoeA) from Halomonas elongata | | Descriptor: | (2~{R})-4-azanyl-2-[[(1~{S})-1-oxidanylethyl]amino]butanoic acid, (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Ectoine hydrolase DoeA | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
6TWL
 
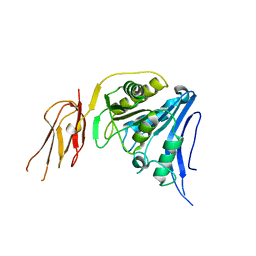 | |
6TI2
 
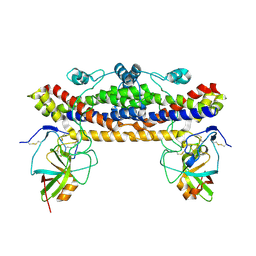 | |
7OJ2
 
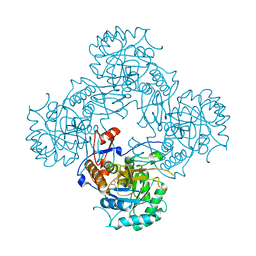 | | Bacillus subtilis IMPDH in complex with Ap4A | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Giammarinaro, P.I, Bange, G. | | Deposit date: | 2021-05-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Nat Microbiol, 7, 2022
|
|
5JRL
 
 | |
5JQF
 
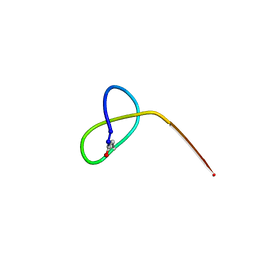 | | Crystal structure of the lasso peptide Sphingopyxin I (SpI) | | Descriptor: | Sphingopyxin I | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-04 | | Release date: | 2016-09-14 | | Last modified: | 2021-06-16 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5J8Q
 
 | |
5JRK
 
 | | Crystal Structure of the Sphingopyxin I Lasso Peptide Isopeptidase SpI-IsoP (SeMet-derived) | | Descriptor: | Dipeptidyl aminopeptidases/acylaminoacyl-peptidases-like protein, beta-D-glucopyranose | | Authors: | Fage, C.D, Hegemann, J.D, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5JAK
 
 | | Crystal structure of the flagellar assembly factor FliW | | Descriptor: | Flagellar assembly factor FliW | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2016-04-12 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6YO9
 
 | |
6Z0W
 
 | |
6YVC
 
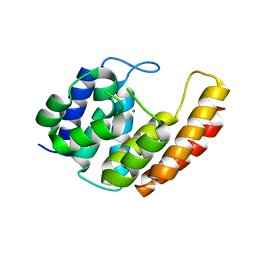 | |
6YXA
 
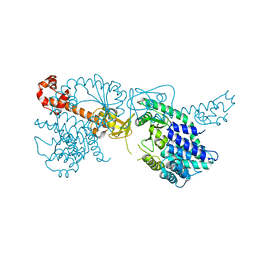 | | Structure of the bifunctional Rel enzyme from B. subtilis | | Descriptor: | GTP pyrophosphokinase, MANGANESE (II) ION | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structural Basis for Regulation of the Opposing (p)ppGpp Synthetase and Hydrolase within the Stringent Response Orchestrator Rel.
Cell Rep, 32, 2020
|
|
6G15
 
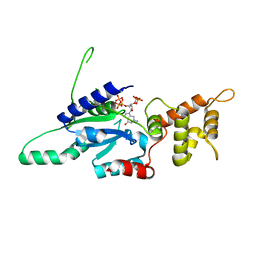 | | Crystal structure of pppGpp bound RbgA from S. aureus | | Descriptor: | Ribosome biogenesis GTPase A, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6GOW
 
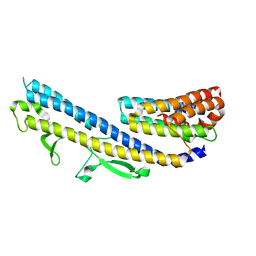 | |
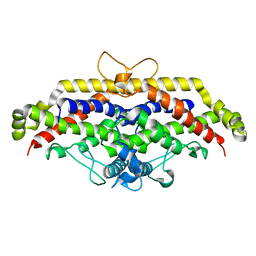
6H3P
 
 | |
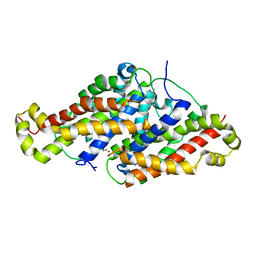
6HJW
 
 | |
6H9H
 
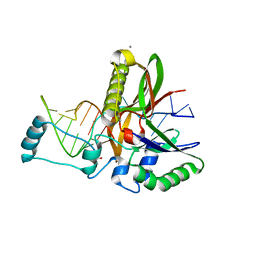 | | Csf5, CRISPR-Cas type IV Cas6 crRNA endonuclease | | Descriptor: | Csf5, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-08-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Nat Microbiol, 4, 2019
|
|
5NJT
 
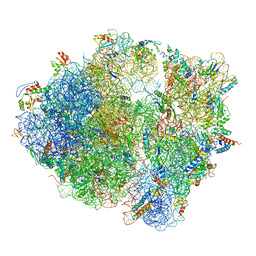 | | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Beckert, B, Abdelshahid, M, Schaefer, H, Steinchen, W, Arenz, S, Berninghausen, O, Beckmann, R, Bange, G, Turgay, K, Wilson, D.N. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.
EMBO J., 36, 2017
|
|
6FPG
 
 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
