4MYD
 
 | | 1.37 Angstrom Crystal Structure of E. Coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) in complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-27 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
2Q8F
 
 | | Structure of pyruvate dehydrogenase kinase isoform 1 | | Descriptor: | POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-26 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
2QLR
 
 | |
2R2N
 
 | | The crystal structure of human kynurenine aminotransferase II in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2007-08-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase II.
J.Biol.Chem., 283, 2008
|
|
2QTW
 
 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5C
 
 | | Aedes Kynurenine Aminotransferase in Complex with Cysteine | | Descriptor: | Kynurenine aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
3EXE
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXI
 
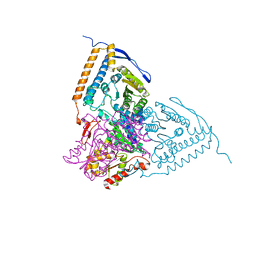 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex with the subunit-binding domain (SBD) of E2p, but SBD cannot be modeled into the electron density | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXH
 
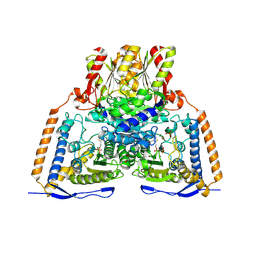 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3FVU
 
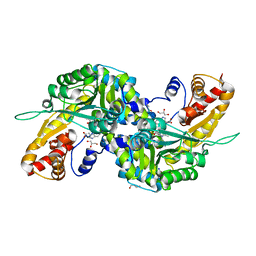 | | Crystal Structure of Human Kynurenine Aminotransferase I in Complex with Indole-3-acetic Acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, GLYCEROL, Kynurenine--oxoglutarate transaminase 1, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3FVS
 
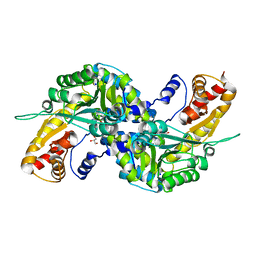 | | Human Kynurenine Aminotransferase I in complex with Glycerol | | Descriptor: | GLYCEROL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3FVX
 
 | | Human kynurenine aminotransferase I in complex with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3EXF
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXG
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3DC1
 
 | | Crystal structure of kynurenine aminotransferase II complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Kynurenine/alpha-aminoadipate aminotransferase mitochondrial | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
Biosci.Rep., 28, 2008
|
|
8WWS
 
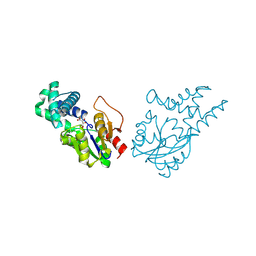 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
8XOY
 
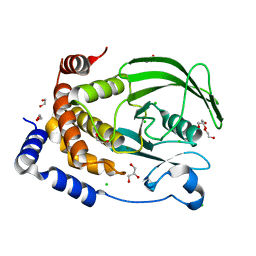 | | The Crystal Structure of PTP1B from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Li, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of PTP1B from Biortus.
To Be Published
|
|
2JXO
 
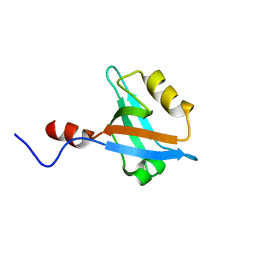 | | Structure of the second PDZ domain of NHERF-1 | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Cheng, H, Li, J, Dai, Z, Bu, Z, Roder, H. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory Interactions between the PDZ2 and C-terminal Domains in the Scaffolding Protein NHERF1
Structure, 17, 2009
|
|
7E6R
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.6 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-23 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6L
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.0 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78037143 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6M
 
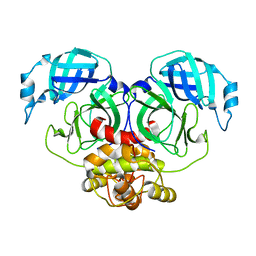 | | Crystal structure of Human coronavirus NL63 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83445024 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7DYS
 
 | |
3RHZ
 
 | |
