4QED
 
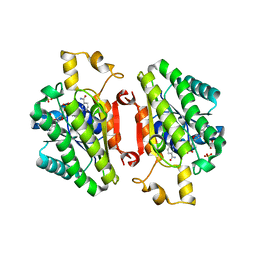 | | ElxO Y152F with NADPH Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4RNM
 
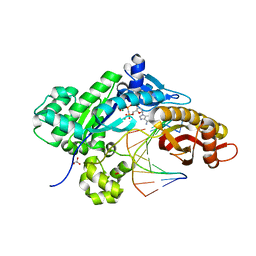 | |
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8UVK
 
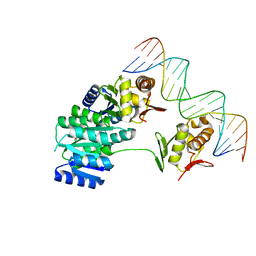 | | CosR DNA bound form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
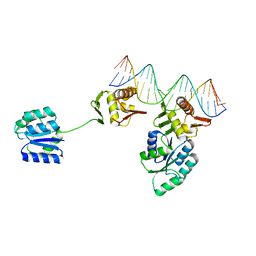 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
5HDX
 
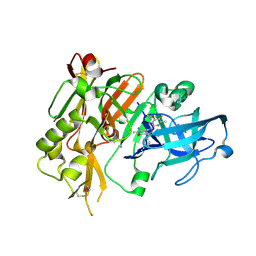 | |
5HE4
 
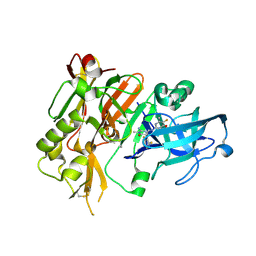 | | BACE-1 in complex with (4aR,7aS)-7a-(2,6-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-3-methyl-4-oxooctahydro-2H-pyrrolo[3,4-d]pyrimidin-2-iminium | | Descriptor: | (2E,4aR,7aS)-7a-(2,6-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-16 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of an Iminoheterocyclic beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE) Inhibitor that Lowers Central A beta in Nonhuman Primates.
J.Med.Chem., 59, 2016
|
|
5HDZ
 
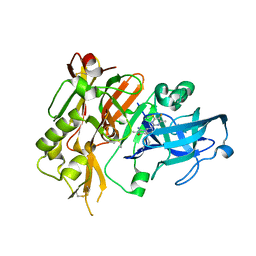 | |
5HE7
 
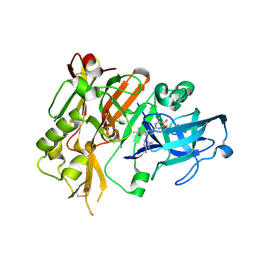 | | BACE-1 in complex with (4aR,7aS)-7a-(2,4-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one | | Descriptor: | (2E,4aR,7aS)-7a-(2,4-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-16 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-Based Design of an Iminoheterocyclic beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE) Inhibitor that Lowers Central A beta in Nonhuman Primates.
J.Med.Chem., 59, 2016
|
|
5HD0
 
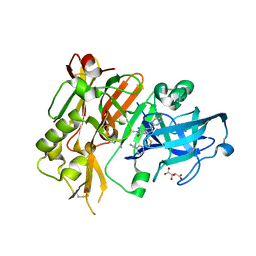 | |
5HDU
 
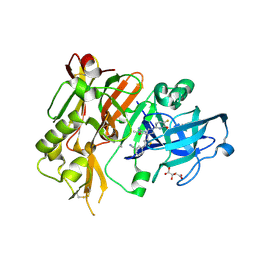 | |
5HDV
 
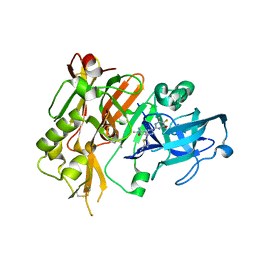 | |
7Y4H
 
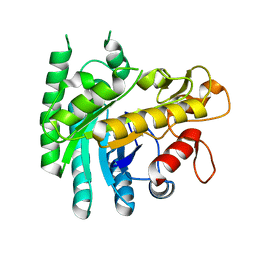 | |
2MJV
 
 | |
8DL9
 
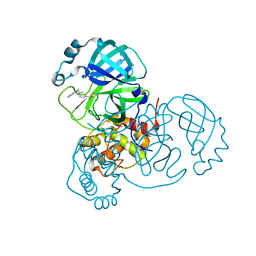 | |
8DLB
 
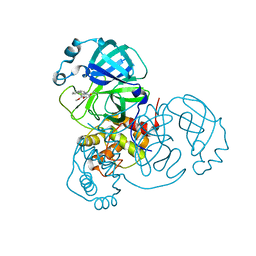 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | Descriptor: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
4DUR
 
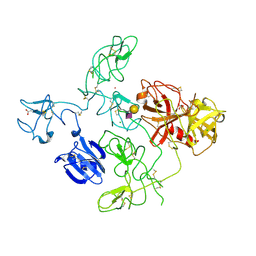 | | The X-ray Crystal Structure of Full-Length type II Human Plasminogen | | Descriptor: | ACETATE ION, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Law, R.H.P, Caradoc-Davies, T, Whisstock, J.C. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The X-ray crystal structure of full-length human plasminogen
Cell Rep, 1, 2012
|
|
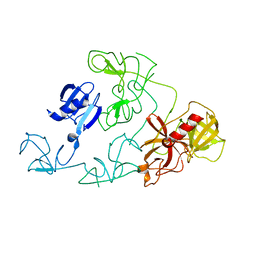
4DUU
 
 | |
4CGR
 
 | |
5ER7
 
 | | Connexin-26 Bound to Calcium | | Descriptor: | CALCIUM ION, Gap junction beta-2 protein | | Authors: | Purdy, M.D, Bennett, B.C, Baker, K.A, Yeager, M.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | An electrostatic mechanism for Ca(2+)-mediated regulation of gap junction channels.
Nat Commun, 7, 2016
|
|
8JSC
 
 | | Structure of the FSP1 protein from Human | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, S.T, Jia, D. | | Deposit date: | 2023-06-19 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | FSP1 oxidizes NADPH to suppress ferroptosis.
Cell Res., 33, 2023
|
|
5ERA
 
 | |
7BYF
 
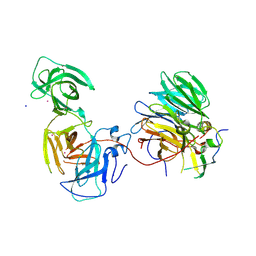 | | The crystal structure of mouse ORF10-Rae1-Nup98 complex | | Descriptor: | 10 protein, MERCURY (II) ION, Peptidase S59 domain-containing protein, ... | | Authors: | Gao, P, Feng, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | Descriptor: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | Authors: | Dong, X, Gong, Z, Hu, Y.F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
