5TSE
 
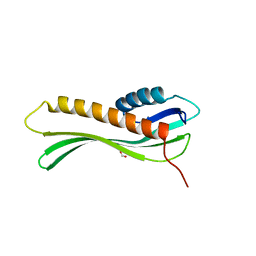 | | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii. | | Descriptor: | FORMIC ACID, LPS-assembly lipoprotein LptE | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii.
To Be Published
|
|
5TF3
 
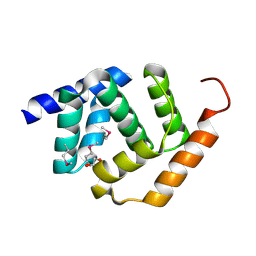 | | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, Putative membrane protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Anderson, W.F, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis
To Be Published
|
|
5TKW
 
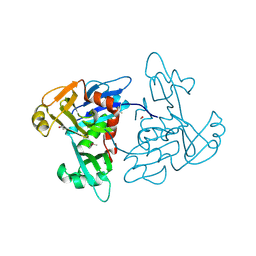 | | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae. | | Descriptor: | FORMIC ACID, Type II secretion system protein L | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae.
To Be Published
|
|
5U4Q
 
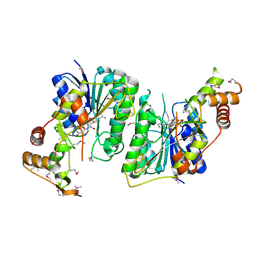 | | 1.5 Angstrom Resolution Crystal Structure of NAD-Dependent Epimerase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-05 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Resolution Crystal Structure of NAD-Dependent Epimerase from Klebsiella pneumoniae in Complex with NAD.
To Be Published
|
|
5TPI
 
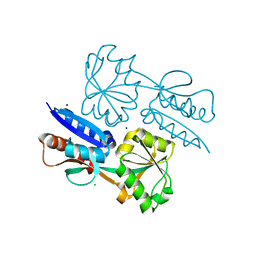 | | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator (LysR family), SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae.
To Be Published
|
|
5TXU
 
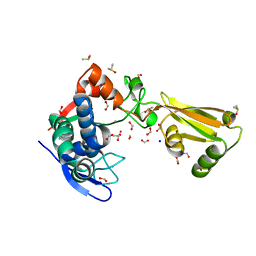 | | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Nocadello, S, Minasov, G, Kiryukhina, O, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation
To Be Published
|
|
5UBU
 
 | | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica. | | Descriptor: | Putative acetamidase/formamidase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica.
To Be Published
|
|
5TU0
 
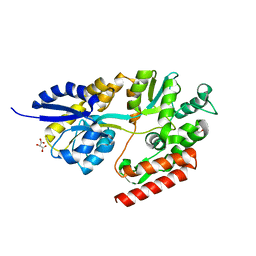 | | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose | | Descriptor: | Lmo2125 protein, TARTRONATE, TRIETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose.
To Be Published
|
|
5U4H
 
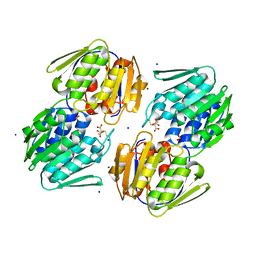 | | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, FORMIC ACID, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid.
To Be Published
|
|
5TTA
 
 | | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein | | Descriptor: | Putative exported protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Cordona-Correa, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein
To Be Published
|
|
3NVS
 
 | | 1.02 Angstrom resolution crystal structure of 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae in complex with shikimate-3-phosphate (partially photolyzed) and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.021 Å) | | Cite: | 1.02 Angstrom Resolution Crystal Structure of 3-Phosphoshikimate 1-Carboxyvinyltransferase from Vibrio cholerae in complex with Shikimate-3-Phosphate (Partially Photolyzed) and Glyphosate
TO BE PUBLISHED
|
|
3OUT
 
 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
3SS6
 
 | | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA acetyltransferase, POTASSIUM ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase
TO BE PUBLISHED
|
|
3VCZ
 
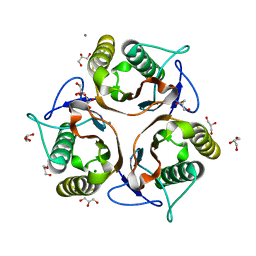 | | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6 | | Descriptor: | CALCIUM ION, Endoribonuclease L-PSP, GLYCEROL, ... | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6
To be Published
|
|
3T4E
 
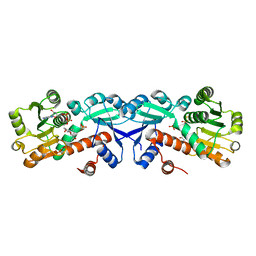 | | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Quinate/shikimate dehydrogenase | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD.
TO BE PUBLISHED
|
|
3T41
 
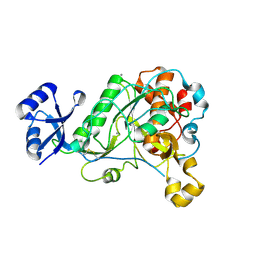 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | Authors: | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3T5P
 
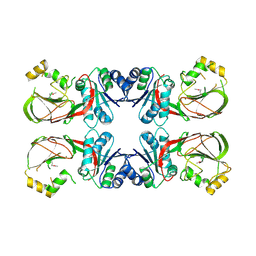 | | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | BmrU protein, MAGNESIUM ION | | Authors: | Hou, J, Zheng, H, Chruszcz, M, Cooper, D.R, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
To be Published
|
|
3UZR
 
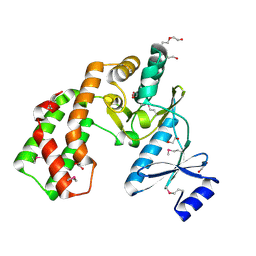 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside phosphotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form
TO BE PUBLISHED
|
|
3T4X
 
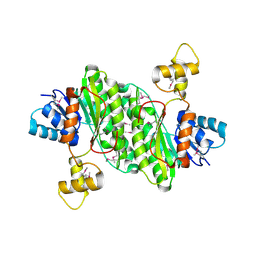 | | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family | | Authors: | Filippova, E.V, Wawrzak, Z, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3URY
 
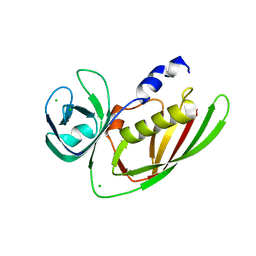 | | Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | CHLORIDE ION, Exotoxin | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
3UGS
 
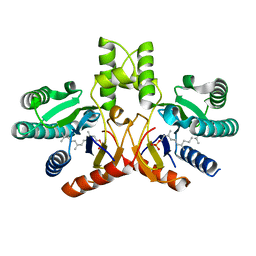 | | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, Undecaprenyl pyrophosphate synthase | | Authors: | Nocek, B, Gu, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni
TO BE PUBLISHED
|
|
3T7B
 
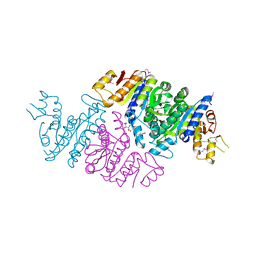 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | Descriptor: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | Authors: | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
3UWQ
 
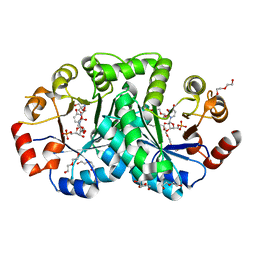 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
3UY4
 
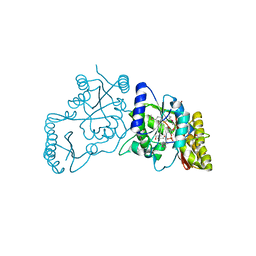 | | Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5
To be Published
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
