5RTX
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000090873 | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008615114 | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUW
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000045014941 | | Descriptor: | 3-{[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]carbamoyl}benzoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000933940912 | | Descriptor: | (1R,5R)-N-methyl-N-(1H-pyrazol-4-yl)bicyclo[3.1.0]hexane-1-carboxamide, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVT
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002582714 | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RG4
 
 | | Crystal Structure of Kemp Eliminase HG3 in unbound state, 277K | | Descriptor: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RSK
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000901381520_N3 | | Descriptor: | 3-[(3-methoxy-1,2-oxazol-5-yl)methyl]-3H-purin-6-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSZ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000004218283 | | Descriptor: | 1-PHENYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTE
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000013283576 | | Descriptor: | 4-(1H-imidazol-2-yl)pyridine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSF
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000026180281 | | Descriptor: | 9-methyl-9H-purine-2,6-diamine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001698894 | | Descriptor: | 3-hydroxy-2-methylbenzoic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSX
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388262 | | Descriptor: | 2-(3-methoxy-4-oxidanyl-phenyl)ethanoic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTD
 
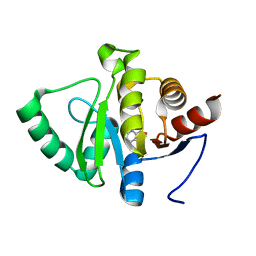 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000157108 | | Descriptor: | 3-HYDROXYPICOLINIC ACID, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTT
 
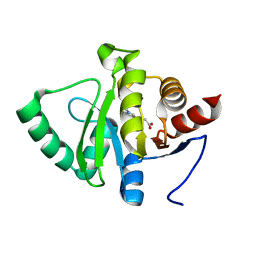 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000873830 | | Descriptor: | 3-(1-methyl-1H-indol-3-yl)propanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUS
 
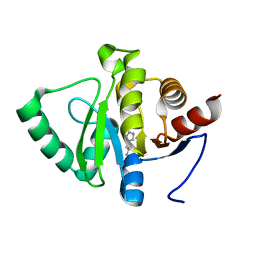 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388081 | | Descriptor: | HISTAMINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU9
 
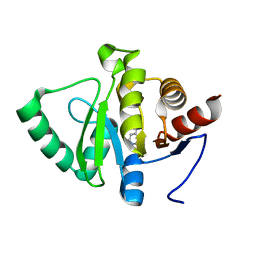 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000165882 | | Descriptor: | 3-AMINOPYRIDINE-4-CARBOXYLIC ACID, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV9
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388150 | | Descriptor: | 4-tert-butylbenzene-1,2-diol, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUN
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000194295 | | Descriptor: | 3-(1H-benzimidazol-1-yl)propanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000013514509 | | Descriptor: | 2-AMINOQUINAZOLIN-4(3H)-ONE, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV5
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008578948 | | Descriptor: | BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000084843283 | | Descriptor: | CHLORZOXAZONE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RGA
 
 | | Crystal Structure of Kemp Eliminase HG3 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGC
 
 | | Crystal Structure of Kemp Eliminase HG3.7 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RSN
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000064576 | | Descriptor: | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RT3
 
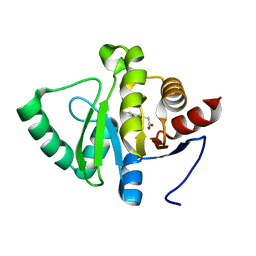 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000039281982 | | Descriptor: | 1-benzothiophen-2-ylacetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
