2YYO
 
 | | Crystal structure of human SPRY domain | | Descriptor: | SPRY domain-containing protein 3 | | Authors: | Kishishita, S, Uchikubo-Kamo, T, Murayama, K, Terada, T, Chen, L, Fu, Z.Q, Chrzas, J, Shirouzu, M, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human SPRY domain
To be Published
|
|
2Z0R
 
 | | Crystal structure of hypothetical protein TTHA0547 | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein TTHA0547, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of hypothetical protein TTHA0547
to be published
|
|
2YZY
 
 | | Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Putative uncharacterized protein TTHA1012 | | Authors: | Ebihara, A, Watanabe, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8
To be Published
|
|
2Z09
 
 | |
3AYT
 
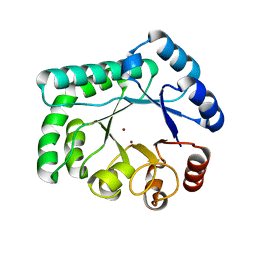 | | TTHB071 protein from Thermus thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
3AYV
 
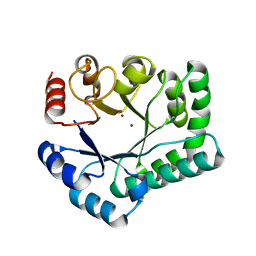 | | TTHB071 protein from Thermus thermophilus HB8 soaking with ZnCl2 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
3AXS
 
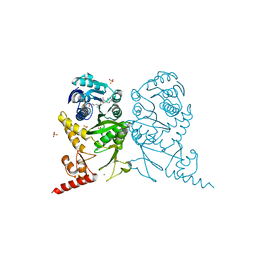 | | Complex structure of tRNA methyltransferase Trm1 from Aquifex aeolicus with sinefungin | | Descriptor: | Probable N(2),N(2)-dimethylguanosine tRNA methyltransferase Trm1, SINEFUNGIN, SULFATE ION, ... | | Authors: | Ihsanawati, Sengoku, T, Yokoyama, S, Bessho, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-04-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure
J.Biol.Chem., 286, 2011
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
2Z1Q
 
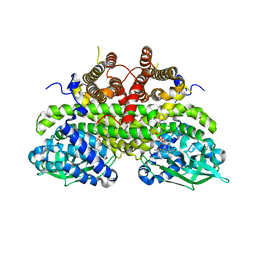 | |
3CPQ
 
 | | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044) | | Descriptor: | 50S ribosomal protein L30e | | Authors: | Jeyakanthan, J, Sarani, R, Mridula, P, Sekar, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044)
To be Published
|
|
3CQ2
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Ebihara, A, Chen, L, Liu, Z.J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein from Thermus Thermophilus HB8
To be Published
|
|
2Z1K
 
 | | Crystal Structure of Ttha1563 from Thermus thermophilus HB8 | | Descriptor: | (Neo)pullulanase, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), PHOSPHATE ION, ... | | Authors: | Niwa, H, Shimada, A, Matsunaga, E, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Ttha1563 from Thermus thermophilus HB8
To be Published
|
|
1UHT
 
 | | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein | | Descriptor: | expressed protein | | Authors: | Tomizawa, T, Inoue, M, Koshiba, S, Hayashi, F, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Shinozaki, K, Seki, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein
To be Published
|
|
2ZTG
 
 | | Crystal structure of Archaeoglobus fulgidus alanyl-tRNA synthetase lacking the C-terminal dimerization domain in complex with Ala-SA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanyl-tRNA synthetase, ZINC ION | | Authors: | Naganuma, M, Sekine, S, Fukunaga, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZUL
 
 | | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Wang, H, Kawazoe, M, Tatsuguchi, A, Naoe, C, Takemoto, C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine
To be Published
|
|
2ZDH
 
 | |
2Z95
 
 | |
1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
2ZP1
 
 | |
2ZR3
 
 | |
2ZV3
 
 | |
2ZDQ
 
 | |
2ZIO
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AlocLys-AMP and PNP | | Descriptor: | 5'-O-[(S)-({(2S)-2-amino-6-[(propoxycarbonyl)amino]hexanoyl}oxy)(hydroxy)phosphoryl]adenosine, IMIDODIPHOSPHORIC ACID, Pyrrolysyl-tRNA synthetase | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
2ZBT
 
 | | Crystal structure of pyridoxine biosynthesis protein from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Pyridoxal biosynthesis lyase pdxS | | Authors: | Manzoku, M, Ebihara, A, Fujimoto, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-29 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of pyridoxine biosynthesis protein from Thermus thermophilus HB8
To be Published
|
|
2ZCW
 
 | | Crystal Structure of TTHA1359, a Transcriptional Regulator, CRP/FNR family from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Transcriptional regulator, FNR/CRP family | | Authors: | Agari, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-13 | | Release date: | 2007-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
