2KAF
 
 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2KU6
 
 | |
2KU5
 
 | |
2KU4
 
 | | Horse prion protein | | Descriptor: | Major prion protein | | Authors: | Perez, D.R, Damberger, F.F, Wuthrich, K. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein.
J.Mol.Biol., 400, 2010
|
|
2KL4
 
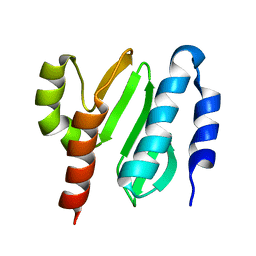 | | NMR structure of the protein NB7804A | | Descriptor: | BH2032 protein | | Authors: | Mohanty, B, Geralt, M, Augustyniak, W, Serrano, P, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein NB7804A from Bacillus Halodurans
To be Published
|
|
2KK2
 
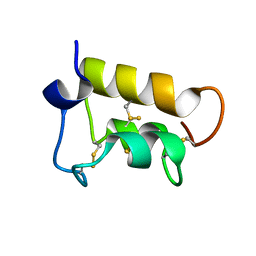 | | NMR solution structure of the pheromone En-A1 from Euplotes nobilii | | Descriptor: | En-A1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antarctic and Arctic populations of the ciliate Euplotes nobilii show common pheromone-mediated cell-cell signaling and cross-mating.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2K5O
 
 | |
2K87
 
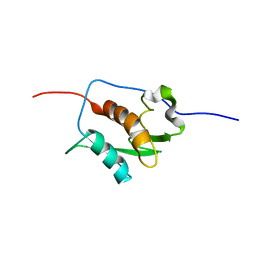 | | NMR STRUCTURE OF A PUTATIVE RNA BINDING PROTEIN (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | Non-structural protein 3 of Replicase polyprotein 1a | | Authors: | Serrano, P, Wuthrich, K, Johnson, M.A, Chatterjee, A, Wilson, I, Pedrini, B.F, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3.
J.Virol., 83, 2009
|
|
2L1S
 
 | | Yp_001336205.1 | | Descriptor: | Uncharacterized protein yohN | | Authors: | Wahab, A, Serrano, P, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-08-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Klebsiella pneumoniae protein YP_001336205
To be Published
|
|
2L1E
 
 | | Mouse prion protein (121-231) containing the substitution F175A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2012-10-24 | | Method: | SOLUTION NMR | | Cite: | Prion Protein mPrP[F175A](121-231): Structure and Stability in Solution.
J.Mol.Biol., 423, 2012
|
|
2L1D
 
 | | Mouse prion protein (121-231) containing the substitution Y169G | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2011-11-09 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L1T
 
 | |
2LQ5
 
 | |
2L6O
 
 | | NMR structure of the protein YP_926445.1 from Shewanella Amazonensis | | Descriptor: | uncharacterized protein YP_926445.1 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Horst, R, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_926445.1 from Shewanella Amazonensis
To be Published
|
|
1MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2KZC
 
 | | Solution NMR structure of the protein YP_510488.1 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the protein YP_510488.1
To be Published
|
|
2L25
 
 | | Np_888769.1 | | Descriptor: | Uncharacterized protein | | Authors: | Wahab, A, Serrano, P, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein NP_888769.1, a phage-related protein in the Bordetella bronchiseptica genome
To be Published
|
|
2L9D
 
 | | Solution structure of the protein YP_546394.1, the first structural representative of the pfam family PF12112 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein YP_546394.1, the first structural representative of the pfam family PF12112
To be Published
|
|
2LLG
 
 | | NMR structure of the protein NP_814968.1 from Enterococcus faecalis | | Descriptor: | Uncharacterized protein | | Authors: | Susac, L, Serrano, P, Geralt, M, Mohanty, B, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_814968.1 from Enterococcus faecalis
To be Published
|
|
4AIT
 
 | |
1ATX
 
 | |
1BF8
 
 | | PERIPLASMIC CHAPERONE FIMC, NMR, 20 STRUCTURES | | Descriptor: | CHAPERONE PROTEIN FIMC | | Authors: | Pellecchia, M, Guntert, P, Glockshuber, R, Wuthrich, K. | | Deposit date: | 1998-05-28 | | Release date: | 1998-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the periplasmic chaperone FimC.
Nat.Struct.Biol., 5, 1998
|
|
3AIT
 
 | |
8FX3
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-GP, showing the structure of the complete active site in its open conformation | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX2
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
