1TMN
 
 | |
6CRO
 
 | |
3LZM
 
 | |
4V45
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
3LVB
 
 | | Crystal structure of the Ferredoxin:NADP+ reductase from maize root at 1.7 angstroms - Test Set Withheld | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues
Biochemistry, 40, 2001
|
|
3F8V
 
 | |
3FA0
 
 | |
3FAD
 
 | |
3F9L
 
 | |
3ORC
 
 | |
1L56
 
 | |
1L60
 
 | |
3DBK
 
 | | Pseudomonas aeruginosa elastase with phosphoramidon | | Descriptor: | CALCIUM ION, Elastase, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | McKay, D.B, Overgaard, M.T. | | Deposit date: | 2008-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Elastase of Pseudomonas aeruginosa Complexed with Phosphoramidon
To be Published
|
|
3FI5
 
 | | Crystal Structure of T4 Lysozyme Mutant R96W | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Lysozyme, ... | | Authors: | Mooers, B.H.M, Matthews, B.W. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C8R
 
 | |
3C7W
 
 | |
3C7Z
 
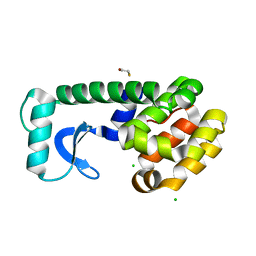 | | T4 lysozyme mutant D89A/R96H at room temperature | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C83
 
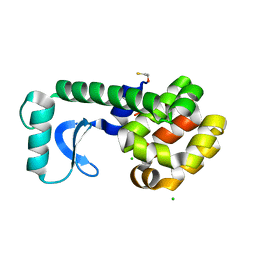 | |
3C81
 
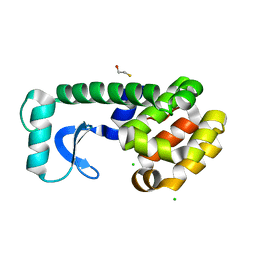 | |
3C8S
 
 | |
3C7Y
 
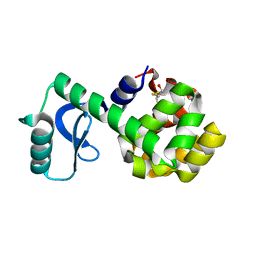 | | Mutant R96A OF T4 lysozyme in wildtype background at 298K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C82
 
 | | Bacteriophage lysozyme T4 lysozyme mutant K85A/R96H | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C8Q
 
 | |
3CDT
 
 | |
3CDV
 
 | |
