5GZ1
 
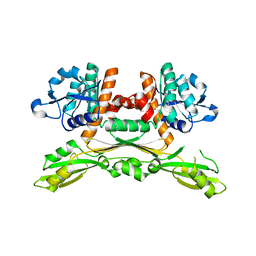 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
8HYE
 
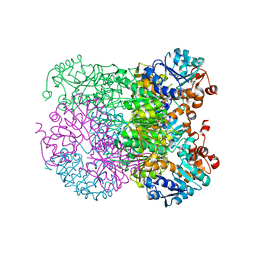 | | Structure of amino acid dehydrogenase-2752 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine dehydrogenase, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
8HYH
 
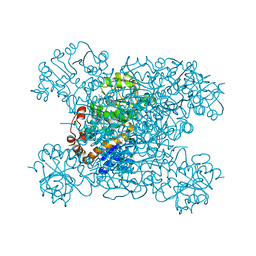 | | Structure of amino acid dehydrogenase3448 | | Descriptor: | 1,2-ETHANEDIOL, Alanine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
8J1G
 
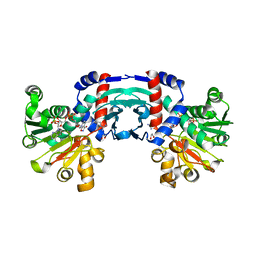 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
8J1C
 
 | |
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WYC
 
 | | Structure of a meso-diaminopimelate dehydrogenase in complex with NADP | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WYB
 
 | | Structure of a meso-diaminopimelate dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FB3
 
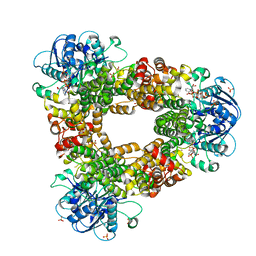 | | Structure of glycerophosphate dehydrogenase in complex with NADPH | | Descriptor: | Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE, ... | | Authors: | Sakuraba, H, Hayashi, J, Yamamoto, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique coenzyme binding mode of hyperthermophilic archaeal sn-glycerol-1-phosphate dehydrogenase from Pyrobaculum calidifontis
Proteins, 84, 2016
|
|
6K9Z
 
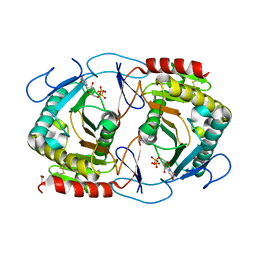 | | STRUCTURE OF URIDYLYLTRANSFERASE MUTANT | | Descriptor: | ACETATE ION, FE (III) ION, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
4XB2
 
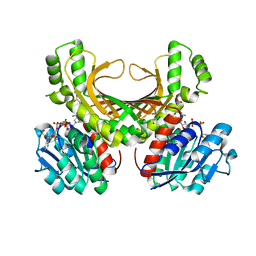 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | Descriptor: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4XB1
 
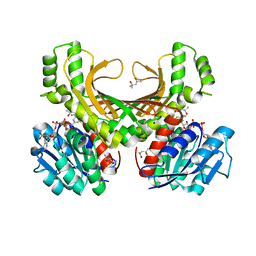 | | Hyperthermophilic archaeal homoserine dehydrogenase in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 319aa long hypothetical homoserine dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4YSV
 
 | |
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3WID
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with NADP | | Descriptor: | Glucose 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WIE
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5B37
 
 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
2D4A
 
 | | Structure of the malate dehydrogenase from Aeropyrum pernix | | Descriptor: | Malate dehydrogenase | | Authors: | Kawakami, R, Sakuraba, H, Tsuge, H, Ohshima, T. | | Deposit date: | 2005-10-12 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Refolding, characterization and crystal structure of (S)-malate dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
Biochim.Biophys.Acta, 1794, 2009
|
|
2DC1
 
 | | Crystal Structure Of L-Aspartate Dehydrogenase From Hyperthermophilic Archaeon Archaeoglobus fulgidus | | Descriptor: | CITRIC ACID, L-aspartate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yoneda, K, Sakuraba, H, Tsuge, H, Ohshima, T. | | Deposit date: | 2005-12-19 | | Release date: | 2006-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of archaeal highly thermostable L-aspartate dehydrogenase/NAD/citrate ternary complex.
Febs J., 274, 2007
|
|
1L2L
 
 | |
5XVH
 
 | | Crystal structure of the NADP+ and tartrate-bound complex of L-serine 3-dehydrogenase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | 6-phosphogluconate dehydrogenase, NAD-binding protein, ACETIC ACID, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2017-06-28 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the NADP+and tartrate-bound complex of L-serine 3-dehydrogenase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Extremophiles, 22, 2018
|
|
2DC0
 
 | | Crystal structure of amidase | | Descriptor: | probable amidase | | Authors: | Ohshima, T, Sakuraba, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Satoh, S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-17 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amidase
To be Published
|
|
6JYG
 
 | | Crystal Structure of L-threonine dehydrogenase from Phytophthora infestans | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CITRATE ANION, L-threonine 3-dehydrogenase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic properties and crystal structure of UDP-galactose 4-epimerase-like l-threonine 3-dehydrogenase from Phytophthora infestans.
Enzyme.Microb.Technol., 140, 2020
|
|
1Y56
 
 | | Crystal structure of L-proline dehydrogenase from P.horikoshii | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tsuge, H, Kawakami, R, Sakuraba, H, Ago, H, Miyano, M, Aki, K, Katunuma, N, Ohshima, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of a novel FAD-, FMN-, and ATP-containing L-proline dehydrogenase complex from Pyrococcus horikoshii
J.Biol.Chem., 280, 2005
|
|
