4XJ7
 
 | |
1SMV
 
 | |
4XER
 
 | | Crystal Structure of C2 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, ACETATE ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XH8
 
 | |
2HTA
 
 | | Crystal Structure of a putative mutarotase (YeaD) from Salmonella typhimurium in orthorhombic form | | Descriptor: | GLYCEROL, Putative enzyme related to aldose 1-epimerase, SULFATE ION | | Authors: | Chittori, S, Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2HTB
 
 | | Crystal Structure of a putative mutarotase (YeaD) from Salmonella typhimurium in monoclinic form | | Descriptor: | Putative enzyme related to aldose 1-epimerase | | Authors: | Chittori, S, Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4R2K
 
 | |
4R2J
 
 | |
4R2L
 
 | |
4R2M
 
 | |
4RYU
 
 | |
4Y5Z
 
 | |
4RYT
 
 | |
1YDV
 
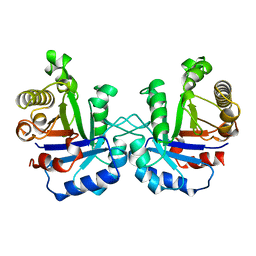 | | TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Velankar, S.S, Murthy, M.R.N. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design.
Structure, 5, 1997
|
|
3PWA
 
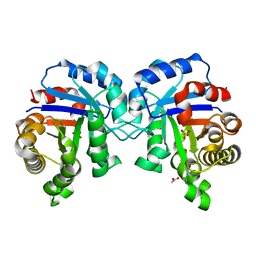 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3PVF
 
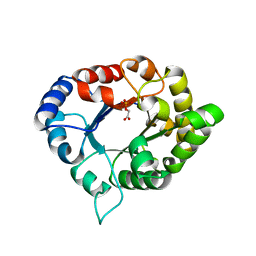 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3PY2
 
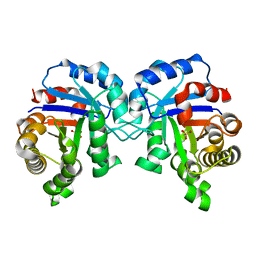 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
2GN0
 
 | |
4ZZ9
 
 | | Crystal structure of T75S mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
4ZRC
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BMX
 
 | | Crystal structure of T75N mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BMW
 
 | | Crystal structure of T75V mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BRB
 
 | | Crystal structure of Q64E mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-30 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BNK
 
 | | Crystal structure of T75C mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BYV
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
