3P2N
 
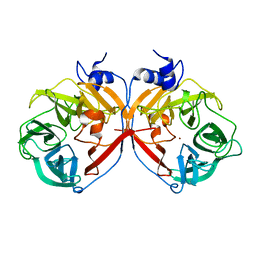 | | Discovery and structural characterization of a new glycoside hydrolase family abundant in coastal waters that was annotated as 'hypothetical protein' | | Descriptor: | 3,6-anhydro-alpha-L-galactosidase, CHLORIDE ION, ZINC ION | | Authors: | Rebuffet, E, Barbeyron, T, Czjzek, M, Michel, G. | | Deposit date: | 2010-10-03 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and structural characterization of a novel glycosidase family of marine origin.
Environ Microbiol, 13, 2011
|
|
3ZPY
 
 | | Crystal structure of the marine PL7 alginate lyase AlyA1 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, SODIUM ION | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
4BE3
 
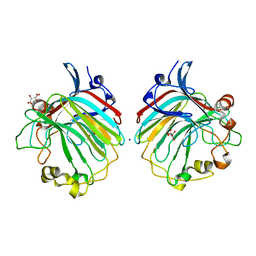 | | crystal structure of the exolytic PL7 alginate lyase AlyA5 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, D(-)-TARTARIC ACID, ... | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
5OPQ
 
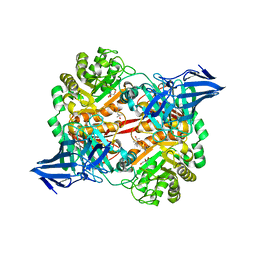 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
4BPZ
 
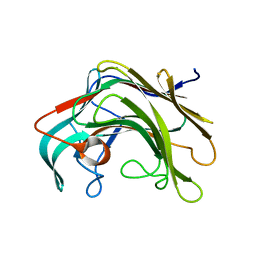 | | Crystal structure of lamA_E269S from Zobellia galactanivorans in complex with a trisaccharide of 1,3-1,4-beta-D-glucan. | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin.
J.Biol.Chem., 289, 2014
|
|
4BOW
 
 | | Crystal structure of LamA_E269S from Z. galactanivorans in complex with laminaritriose and laminaritetraose | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
4BQ1
 
 | | Crystal structure of of LamAcat from Zobellia galactanivorans | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-05-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Beta Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
4CTE
 
 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S in complex with a thio-oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, 1-thio-beta-D-glucopyranose-(1-3)-1-thio-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CIT
 
 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-10-08 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
6GL0
 
 | | Structure of ZgEngAGH5_4 in complex with a cellotriose | | Descriptor: | Endoglucanase, family GH5, MAGNESIUM ION, ... | | Authors: | Dorival, J, Ruppert, S, Gunnoo, M, Orlowski, A, Chapelais, M, Dabin, J, Labourel, A, Thompson, D, Michel, G, Czjzek, M, Genicot, S. | | Deposit date: | 2018-05-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The laterally acquired GH5ZgEngAGH5_4from the marine bacteriumZobellia galactanivoransis dedicated to hemicellulose hydrolysis.
Biochem. J., 475, 2018
|
|
6GL2
 
 | | Structure of ZgEngAGH5_4 wild type at 1.2 Angstrom resolution | | Descriptor: | Endoglucanase, family GH5, IMIDAZOLE | | Authors: | Dorival, J, Ruppert, S, Gunnoo, M, Orlowski, A, Chapelais, M, Dabin, J, Labourel, A, Thompson, D, Michel, G, Czjzek, M, Genicot, S. | | Deposit date: | 2018-05-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The laterally acquired GH5ZgEngAGH5_4from the marine bacteriumZobellia galactanivoransis dedicated to hemicellulose hydrolysis.
Biochem. J., 475, 2018
|
|
5OCQ
 
 | | Crystal structure of the complex of the kappa-carrageenase from Pseudoalteromonas carrageenovora with an oligotetrasaccharide of kappa-carrageenan | | Descriptor: | 3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CITRIC ACID, Kappa-carrageenase | | Authors: | Czjzek, M, Leroux, C, Bernard, T, Matard-Mann, M, Jeudy, A, Michel, G. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
5OCR
 
 | | Crystal structure of the kappa-carrageenase zobellia_236 from Zobellia galactanivorans | | Descriptor: | GLYCEROL, Kappa-carrageenase, MAGNESIUM ION | | Authors: | Czjzek, M, Matard-Mann, M, Michel, G, Jeudy, A, Larocque, R. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
4A15
 
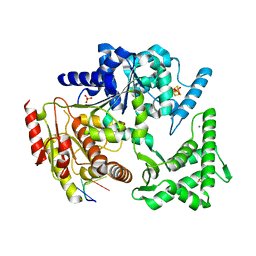 | | Crystal structure of an XPD DNA complex | | Descriptor: | 5'-D(*DTP*AP*CP*GP)-3', ATP-DEPENDENT DNA HELICASE TA0057, CALCIUM ION, ... | | Authors: | Kuper, J, Wolski, S.C, Michels, G, Kisker, C. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and Structural Studies of the Nucleotide Excision Repair Helicase Xpd Suggest a Polarity for DNA Translocation.
Embo J., 31, 2011
|
|
1NYT
 
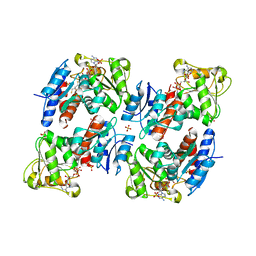 | | SHIKIMATE DEHYDROGENASE AroE COMPLEXED WITH NADP+ | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Roszak, A.W, Lapthorn, A.J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities.
J.Biol.Chem., 278, 2003
|
|
8H2A
 
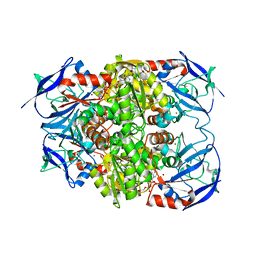 | |
8H2B
 
 | | Crystal structure of alcohol dehydrogenase from Zobellia galactanivorans | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Brott, S, Bornscheuer, U.T, Nam, K.H. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique alcohol dehydrogenases involved in algal sugar utilization by marine bacteria
Appl.Microbiol.Biotechnol., 107, 2023
|
|
7BLY
 
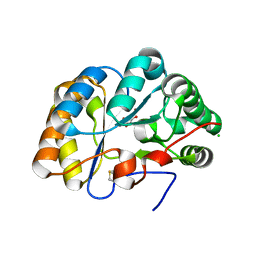 | | Structure of the chitin deacetylase AngCDA from Aspergillus niger | | Descriptor: | Aspergillus niger contig An12c0130, genomic contig, CHLORIDE ION, ... | | Authors: | Roret, T, Bonin, M, Hembach, L, Moerschbacher, B.M. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | In silico and in vitro analysis of an Aspergillus niger chitin deacetylase to decipher its subsite sugar preferences.
J.Biol.Chem., 297, 2021
|
|
7OQD
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose, Arylsulfatase, CALCIUM ION | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2021-06-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
4U6D
 
 | | Zg3615, a family 117 glycoside hydrolase in complex with beta-3,6-anhydro-L-galactose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-beta-L-galactopyranose, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U6B
 
 | | Zg3597, a family 117 glycoside hydrolase, produced by the marine bacterium Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6HPD
 
 | | The structure of a beta-glucuronidase from glycoside hydrolase family 2 | | Descriptor: | BROMIDE ION, Beta-galactosidase (GH2), MAGNESIUM ION | | Authors: | Robb, C.S, Gerlach, N, Reisky, L, Bornshoeru, U, Hehemann, J.H. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
7ALL
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT4683-S1_4) | | Descriptor: | Arylsulfatase, CALCIUM ION, IODIDE ION, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Martens, E.C, Basle, A, Cartmell, A. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7AN1
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | Arylsulfatase, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
