4HZ3
 
 | | MthK pore crystallized in presence of TBSb | | Descriptor: | Calcium-gated potassium channel mthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The voltage-dependent gate in MthK potassium channels is located at the selectivity filter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3RMK
 
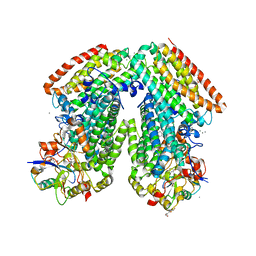 | | Toluene 4 monooxygenase H with 4-bromophenol | | Descriptor: | 4-BROMOPHENOL, CALCIUM ION, FE (III) ION, ... | | Authors: | Bailey, L.J, McCoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3C9Q
 
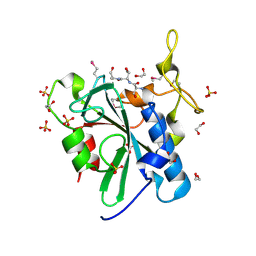 | | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide.
To be Published
|
|
2APJ
 
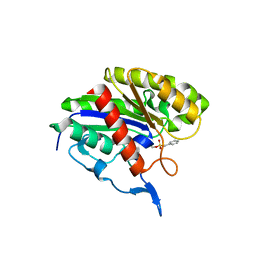 | | X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution | | Descriptor: | Putative Esterase | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure at 1.6 Angstroms resolution of the protein product of the At4g34215 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2IFU
 
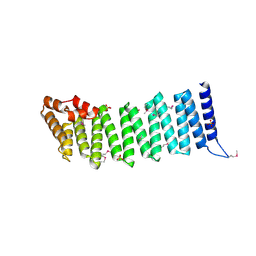 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
2I3C
 
 | | Crystal Structure of an Aspartoacylase from Homo Sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | Descriptor: | hypothetical protein | | Authors: | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-11 | | Release date: | 2006-05-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
2I2O
 
 | | Crystal Structure of an eIF4G-like Protein from Danio rerio | | Descriptor: | NICKEL (II) ION, eIF4G-like protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of an eIF4G-like protein from Danio rerio.
Proteins, 78, 2010
|
|
2NXF
 
 | | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393 | | Descriptor: | ETHANOL, PHOSPHATE ION, Putative dimetal phosphatase, ... | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393
To be Published
|
|
2QU1
 
 | | Crystal Structure of a Cyclized GFP Variant | | Descriptor: | CALCIUM ION, Green fluorescent protein | | Authors: | Bailey, L.J, McCoy, J.G, Bitto, E, Bingman, C.A, Fox, B.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Cyclized GFP Variant.
To be Published
|
|
1Z90
 
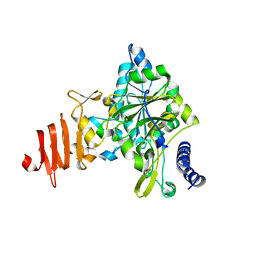 | | X-ray structure of gene product from arabidopsis thaliana at3g03250, a putative UDP-glucose pyrophosphorylase | | Descriptor: | AT3g03250 protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
4LCU
 
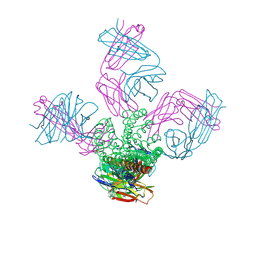 | | Structure of KcsA with E118A mutation | | Descriptor: | DIACYL GLYCEROL, Fab heavy chain, Fab light chain, ... | | Authors: | Nimigean, C.M, Posson, D.J, McCoy, J.M. | | Deposit date: | 2013-06-23 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Molecular interactions involved in proton-dependent gating in KcsA potassium channels.
J.Gen.Physiol., 142, 2013
|
|
4LBE
 
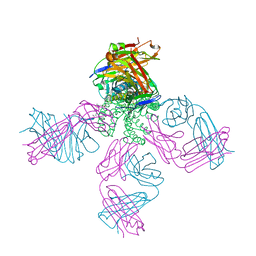 | | Structure of KcsA with R122A mutation | | Descriptor: | DIACYL GLYCEROL, Fab heavy chain, Fab light chain, ... | | Authors: | Nimigean, C.M, Posson, D.J, McCoy, J.M. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Molecular interactions involved in proton-dependent gating in KcsA potassium channels.
J.Gen.Physiol., 142, 2013
|
|
2G09
 
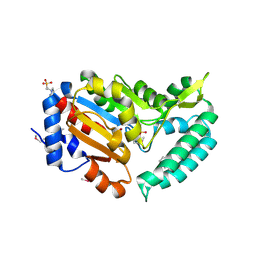 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product complex | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G08
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product-transition complex analog with Aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
4W79
 
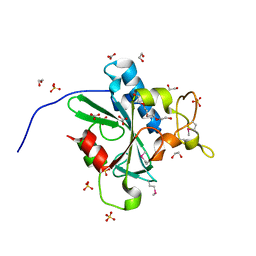 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
8OM2
 
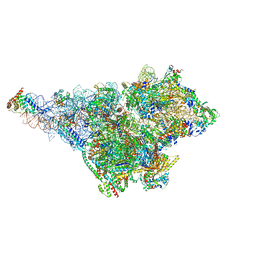 | | Small subunit of yeast mitochondrial ribosome in complex with METTL17/Rsm22. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM4
 
 | | Small subunit of yeast mitochondrial ribosome. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM3
 
 | | Small subunit of yeast mitochondrial ribosome in complex with IF3/Aim23. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
4N7X
 
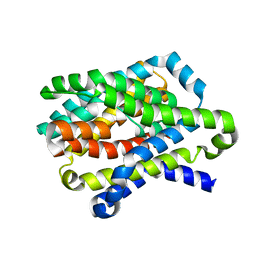 | |
3DKV
 
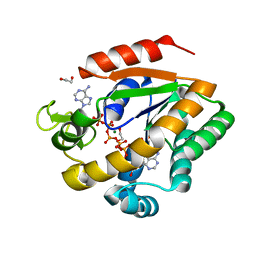 | | Crystal structure of adenylate kinase variant AKlse1 | | Descriptor: | 1,2-ETHANEDIOL, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, Bitto, E.B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of adenylate kinase variant AKlse1.
To be Published
|
|
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
3RI7
 
 | | Toluene 4 monooxygenase HD Mutant G103L | | Descriptor: | ACETATE ION, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Bailey, L.J, Elsen, N.L, Fox, B.G. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3T3P
 
 | | A Novel High Affinity Integrin alphaIIbbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ from the beta3 MIDAS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design of a High-Affinity Platelet Integrin alphaIIbbeta3 Receptor Antagonist That Disrupts Mg2+ Binding to the MIDAS
Sci Transl Med, 4, 2012
|
|
3Q2A
 
 | | Toluene 4 monooxygenase HD complex with inhibitor p-aminobenzoate | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-AMINOBENZOIC ACID, FE (III) ION, ... | | Authors: | Acheson, J.F, Bailey, L.J, Fox, B.G. | | Deposit date: | 2010-12-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
