2L83
 
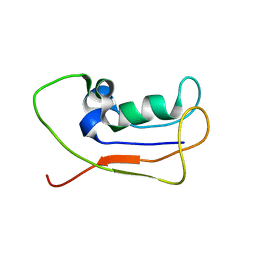 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
2NAS
 
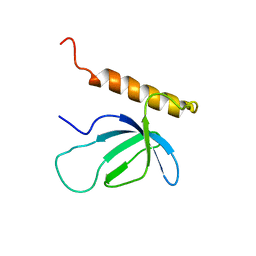 | |
2OIG
 
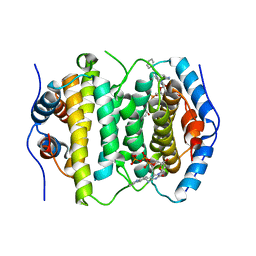 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | Descriptor: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
6ICV
 
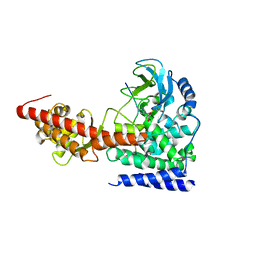 | | Structure of SETD3 bound to SAH and unmodified actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
5GO0
 
 | |
6ICT
 
 | | Structure of SETD3 bound to SAH and methylated actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
7D1L
 
 | | complex structure of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, Uncharacterized protein | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7D2Y
 
 | | complex of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, RRM2, SULFATE ION | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
5YBJ
 
 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBV
 
 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBU
 
 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
7CNF
 
 | | Solution strcture of HsTFIIS LW domain | | Descriptor: | Transcription elongation factor A protein 1 | | Authors: | Gao, J, Liao, S. | | Deposit date: | 2020-07-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of HsTFIIS LW domain
To Be Published
|
|
5ZN9
 
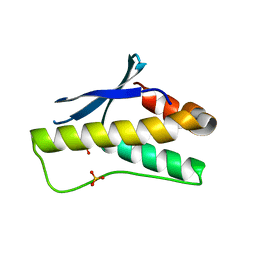 | | Crystal structure of PX domain | | Descriptor: | SULFATE ION, Sorting nexin-27 | | Authors: | Li, Y, Zhu, Z, Li, F, Liao, S, Xu, C. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Crystal structure of PX domain
To Be Published
|
|
6IEV
 
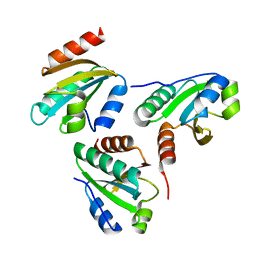 | | Crystal structure of a designed protein | | Descriptor: | Designed protein | | Authors: | Han, M, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Protein Sci., 28, 2019
|
|
6IF4
 
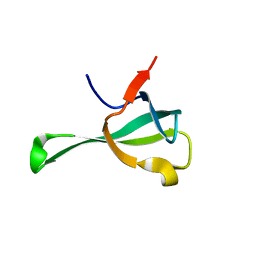 | | Crystal structure of Tbtudor | | Descriptor: | Histone acetyltransferase | | Authors: | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | Deposit date: | 2018-09-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
6K0X
 
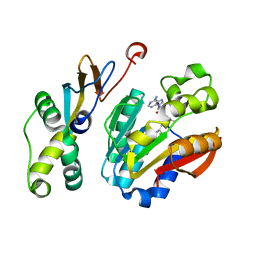 | |
6K6U
 
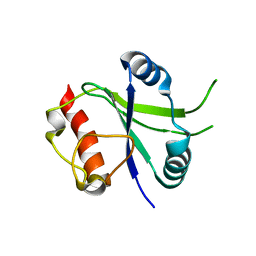 | |
6LBG
 
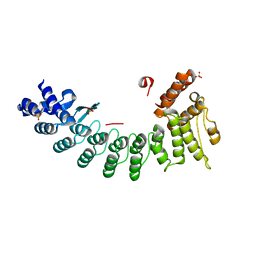 | | Structure of OR51B2 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Olfactory receptor 51B2, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LE6
 
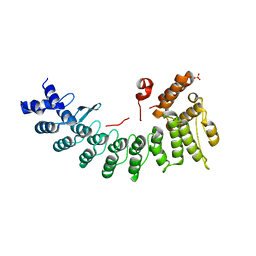 | | Structure of LNLPTQGRAR bound FEM1C | | Descriptor: | Protein fem-1 homolog C,10-mer peptide, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
7CHI
 
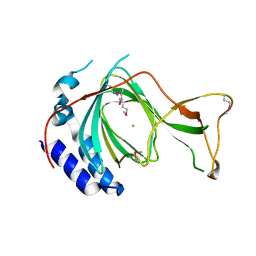 | | crystal structure of pco5 | | Descriptor: | FE (III) ION, Plant cysteine oxidase 5 | | Authors: | Guo, Q, Xu, C, Liao, S. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7CXZ
 
 | | crystal structure of pco2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, NICKEL (II) ION, ... | | Authors: | Guo, Q, Xu, C, Liao, S, Chen, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
7DGY
 
 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7CHJ
 
 | | crystal structure of pco4 | | Descriptor: | CITRIC ACID, FE (III) ION, Plant cysteine oxidase 4 | | Authors: | Guo, Q, Xu, C, Liao, S. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
