8GZX
 
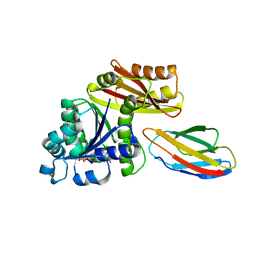 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
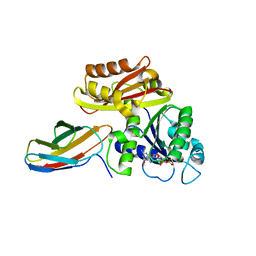 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
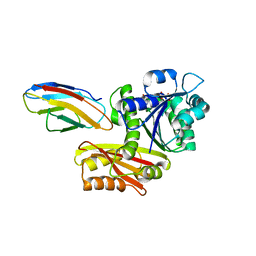 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8H1O
 
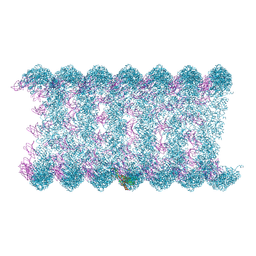 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8IIZ
 
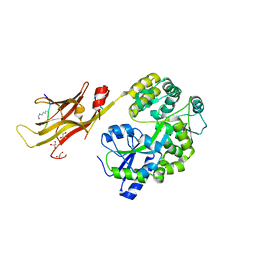 | |
8IIY
 
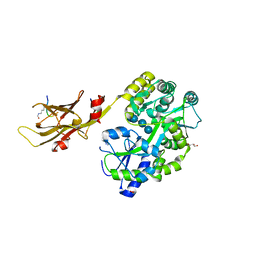 | |
8IJ0
 
 | |
6A7K
 
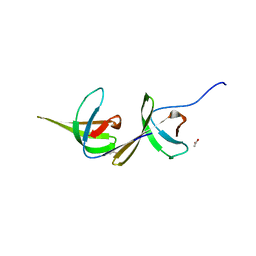 | | X-ray structure of NdhS from T. elongatus | | Descriptor: | ACETIC ACID, Tlr0636 protein | | Authors: | Umeno, K, Misumi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
6LF1
 
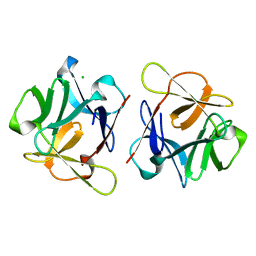 | | SeviL, a GM1b/asialo-GM1 binding lectin | | Descriptor: | CHLORIDE ION, SeviL | | Authors: | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
6LF2
 
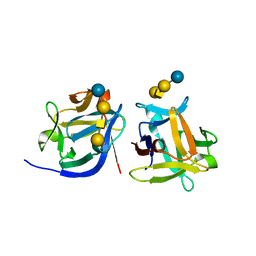 | | SeviL bound to asialo-GM1 saccharide | | Descriptor: | SeviL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
4TLE
 
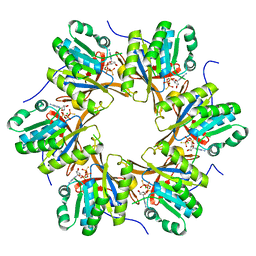 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
4TLC
 
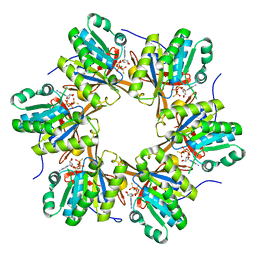 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
4TL7
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Circadian clock protein kinase KaiC, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TLA
 
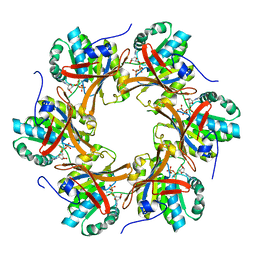 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Circadian clock protein kinase KaiC, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TLB
 
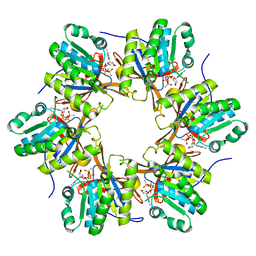 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TL9
 
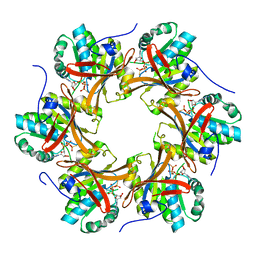 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TL8
 
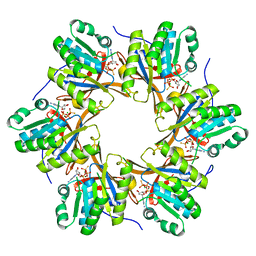 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
4TL6
 
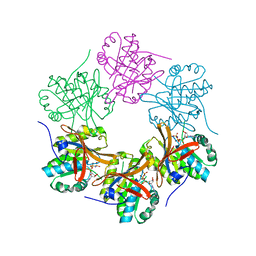 | | Crystal structure of N-terminal domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TLD
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
5Z9C
 
 | |
6IK8
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with beta-1,6-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IK5
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IK7
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with beta-1,3-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IK6
 
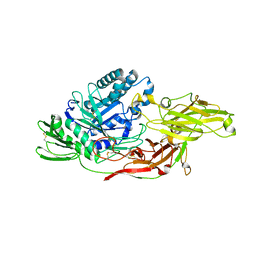 | | Crystal structure of Tomato beta-galactosidase (TBG) 4 with beta-1,4-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6HUM
 
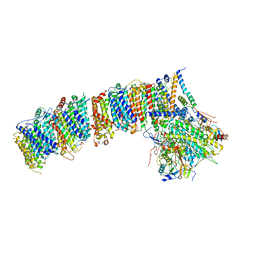 | | Structure of the photosynthetic complex I from Thermosynechococcus elongatus | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, IRON/SULFUR CLUSTER, ... | | Authors: | Schuller, J.M, Schuller, S.K, Kurisu, G, Engel, B.D, Nowaczyk, M.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
