1RFU
 
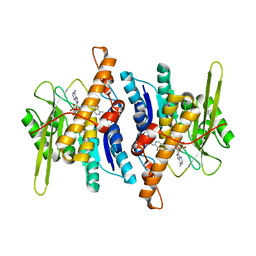 | | Crystal structure of pyridoxal kinase complexed with ADP and PLP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, ZINC ION, ... | | Authors: | Liang, D.-C, Jiang, T, Li, M.-H. | | Deposit date: | 2003-11-10 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes in the reaction of pyridoxal kinase
J.BIOL.CHEM., 279, 2004
|
|
1RFT
 
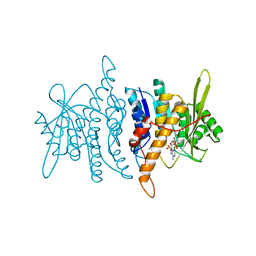 | | Crystal structure of pyridoxal kinase complexed with AMP-PCP and pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Liang, D.-C, Jiang, T, Li, M.-H. | | Deposit date: | 2003-11-10 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes in the reaction of pyridoxal kinase
J.BIOL.CHEM., 279, 2004
|
|
1RFV
 
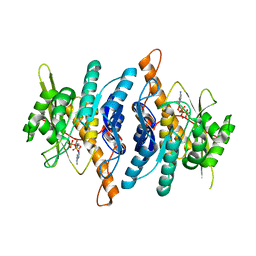 | |
1LHP
 
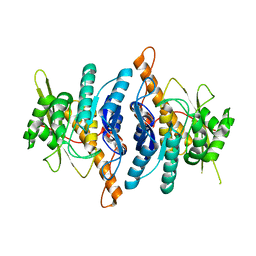 | |
1LHR
 
 | | Crystal Structure of Pyridoxal Kinase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, POTASSIUM ION, Pyridoxal kinase, ... | | Authors: | Liang, D.C, Jiang, T, Li, M.H. | | Deposit date: | 2002-04-17 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of brain pyridoxal kinase, a novel member of the ribokinase superfamily
J.BIOL.CHEM., 277, 2002
|
|
1LIA
 
 | |
1JR9
 
 | | Crystal Structure of manganese superoxide dismutases from Bacillus halodenitrificans | | Descriptor: | MANGANESE (II) ION, ZINC ION, manganese superoxide dismutase | | Authors: | Liao, J, Liu, M.Y, Chang, T, Li, M, LeGall, J, Gui, L.L, Zhang, J.P, Jiang, T, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-08-13 | | Release date: | 2002-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of manganese superoxide dismutase from Bacillus halodenitrificans, a component of the so-called "green protein".
J.Struct.Biol., 139, 2002
|
|
8KBW
 
 | |
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | Descriptor: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
4XPJ
 
 | | Crystal structure of Nerve growth factor in complex with lysophosphatidylinositol | | Descriptor: | (2R)-2-hydroxy-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl tridecanoate, Beta-nerve growth factor | | Authors: | Sun, H.L, Jiang, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of nerve growth factor in complex with lysophosphatidylinositol
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4YN7
 
 | | Non-oxidized YfiR | | Descriptor: | SULFATE ION, YfiR | | Authors: | Yang, X, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YNA
 
 | | Oxidized YfiR | | Descriptor: | SULFATE ION, YfiR | | Authors: | Xu, M, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YN9
 
 | | YfiR mutant-C110S | | Descriptor: | SULFATE ION, YfiR | | Authors: | Xu, M, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
1YGK
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | Pyridoxal kinase, R-ROSCOVITINE | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
1YHJ
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | (2R)-2-{[6-(BENZYLOXY)-9-ISOPROPYL-9H-PURIN-2-YL]AMINO}BUTAN-1-OL, Pyridoxal Kinase | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-09 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives.
J.Biol.Chem., 280, 2005
|
|
1YGJ
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | (2R)-2-({6-[BENZYL(METHYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)BUTAN-1-OL, Pyridoxal kinase | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
4FZX
 
 | | Exonuclease X in complex with 3' overhanging duplex DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*TP*CP*CP*AP*CP*AP*AP*TP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, ... | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
4FZY
 
 | | Exonuclease X in complex with 12bp blunt-ended dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*GP*AP*TP*TP*CP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*GP*AP*AP*TP*CP*TP*AP*CP*A)-3'), Exodeoxyribonuclease 10, ... | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
7YN9
 
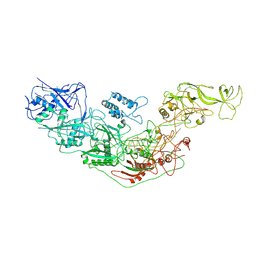 | | Cryo-EM structure of Cas7-11-crRNA binary complex | | Descriptor: | CRISPR-associated RAMP family protein, crRNA | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNC
 
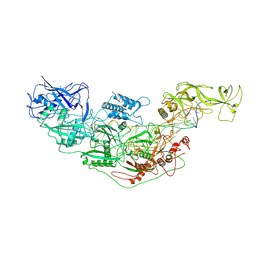 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-3 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-3 (28-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNA
 
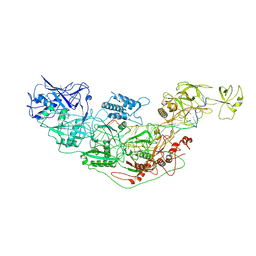 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-1 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-1 (25-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNB
 
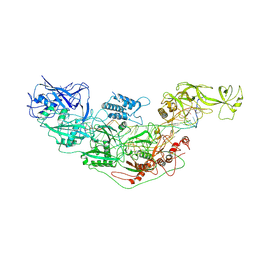 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-2 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-2 (28-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YND
 
 | | Cryo-EM structure of Cas7-11-crRNA-Csx29 ternary complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
4FZZ
 
 | | Exonuclease X in complex with 5' overhanging duplex DNA | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, SODIUM ION | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
