8WCI
 
 | | Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae | | Descriptor: | CARDIOLIPIN, N,N-dimethyl-4-(5-methyl-1H-benzimidazol-2-yl)aniline, SODIUM ION, ... | | Authors: | Suzuki, K, Mikuriya, S, Adachi, N, Kawasaki, M, Senda, T, Moriya, T, Murata, T. | | Deposit date: | 2023-09-12 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Novel Inhibitor of Na+-Transporting V-ATPase Suppresses VRE Colonization in Mice and Reveals the High-Resolution Structure of the Na+ Transport Pathway
To Be Published
|
|
5MXB
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin | | Descriptor: | Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, SODIUM ION, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
5MXW
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
3AN3
 
 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist MO3S | | Descriptor: | (2S)-2-benzyl-3-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}phenyl)propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Waku, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Phenylpropanoic Acid-Type PPAR gamma-Selective Agonists: Discovery of Reversed Stereochemistry-Activity Relationship
J.Med.Chem., 54, 2011
|
|
3AN4
 
 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist MO4R | | Descriptor: | (2R)-2-benzyl-3-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}phenyl)propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Waku, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Phenylpropanoic Acid-Type PPAR gamma-Selective Agonists: Discovery of Reversed Stereochemistry-Activity Relationship
J.Med.Chem., 54, 2011
|
|
5GVI
 
 | | Zebrafish USP30 in complex with Lys6-linked diubiquitin | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, ubiquitin | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30
Nat. Struct. Mol. Biol., 24, 2017
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
6LHM
 
 | | Structure of human PYCR2 | | Descriptor: | Pyrroline-5-carboxylate reductase 2 | | Authors: | Baburajendran, N. | | Deposit date: | 2019-12-09 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Loss of PYCR2 Causes Neurodegeneration by Increasing Cerebral Glycine Levels via SHMT2.
Neuron, 107, 2020
|
|
7DNN
 
 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNM
 
 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DRE
 
 | | Cryo-EM structure of DfgA-B at 2.54 angstrom resolution | | Descriptor: | DfgB, Sugar phosphate isomerase/epimerase | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRD
 
 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DVE
 
 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EXZ
 
 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
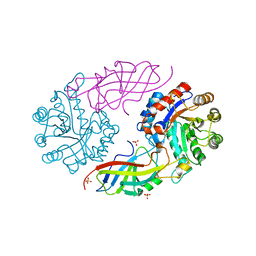 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
4PSB
 
 | |
4Q0K
 
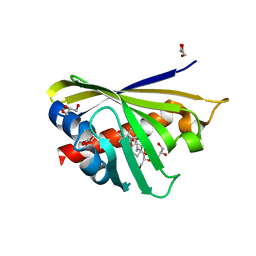 | | Crystal Structure of Phytohormone Binding Protein from Medicago truncatula in complex with gibberellic acid (GA3) | | Descriptor: | GIBBERELLIN A3, GLYCEROL, PHYTOHORMONE BINDING PROTEIN MTPHBP | | Authors: | Ciesielska, A, Barciszewski, J, Ruszkowski, M, Jaskolski, M, Sikorski, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Specific binding of gibberellic acid by Cytokinin-Specific Binding Proteins: a new aspect of plant hormone-binding proteins with the PR-10 fold.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
3C0V
 
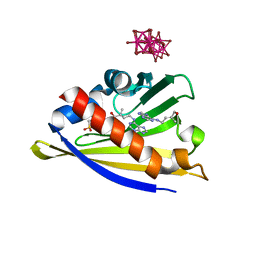 | | Crystal structure of cytokinin-specific binding protein in complex with cytokinin and Ta6Br12 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytokinin-specific binding protein, ... | | Authors: | Pasternak, O, Bujacz, A, Biesiadka, J, Bujacz, G, Sikorski, M, Jaskolski, M. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MAD phasing using the (Ta(6)Br(12))(2+) cluster: a retrospective study
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E85
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Diphenylurea | | Descriptor: | 1,3-DIPHENYLUREA, PR10.2B, SODIUM ION | | Authors: | Fernandes, H.C, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein.
Febs J., 276, 2009
|
|
4JHG
 
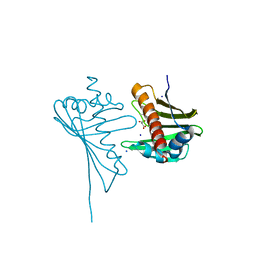 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with trans-zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, MALONATE ION, MtN13 protein, ... | | Authors: | Ruszkowski, M, Tusnio, K, Ciesielska, A, Brzezinski, K, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHI
 
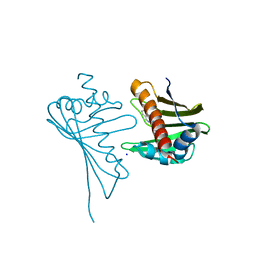 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-benzyladenine | | Descriptor: | MtN13 protein, N-BENZYL-9H-PURIN-6-AMINE, SODIUM ION | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHH
 
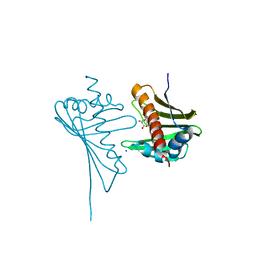 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with kinetin | | Descriptor: | MALONATE ION, MtN13 protein, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GY9
 
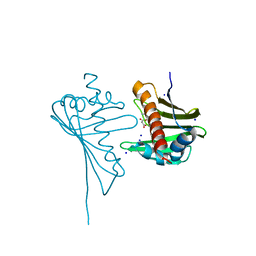 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5I8F
 
 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
