5LIY
 
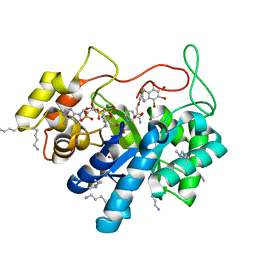 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK204 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIX
 
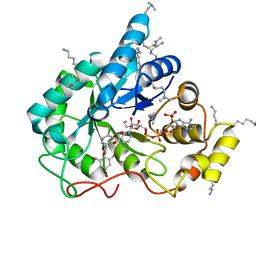 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK184 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIU
 
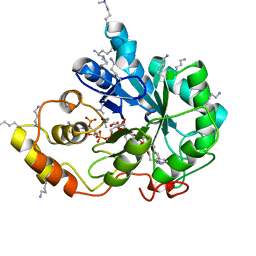 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIW
 
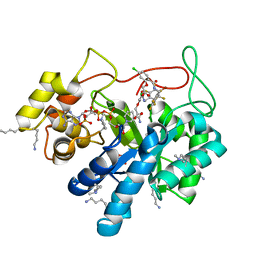 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK319 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
6F8O
 
 | | AKR1B1 at 3.45 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F81
 
 | | AKR1B1 at 0.75 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F82
 
 | | AKR1B1 at 1.65 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F7R
 
 | | AKR1B1 at 0.03 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F84
 
 | | AKR1B1 at 2.55 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
4XZN
 
 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
1PIW
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1PS0
 
 | | Crystal Structure of the NADP(H)-Dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-06-20 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1Q1N
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-07-22 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
7B0U
 
 | | Stressosome complex from Listeria innocua | | Descriptor: | RsbR protein, RsbS protein | | Authors: | Miksys, A, Fu, L, Madej, M.G, Ziegler, C. | | Deposit date: | 2020-11-22 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular insights into intra-complex signal transmission during stressosome activation.
Commun Biol, 5, 2022
|
|
6Z0W
 
 | |
