7B3C
 
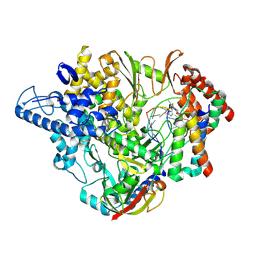 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -4 (structure 2) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
9BZ0
 
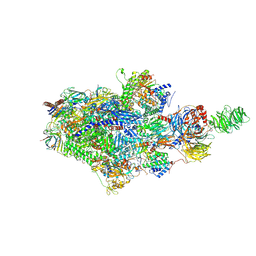 | | Structure of an STK19-containing TC-NER complex | | Descriptor: | DET1- and DDB1-associated protein 1, DNA (35-MER), DNA (49-MER), ... | | Authors: | Mevissen, T.E.T, Kuemmecke, M, Farnung, L, Walter, J.C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | STK19 positions TFIIH for cell-free transcription-coupled DNA repair.
Cell, 187, 2024
|
|
9EGY
 
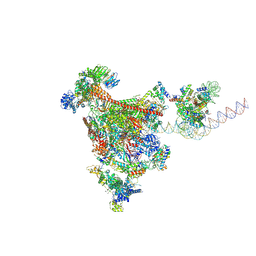 | | RNA polymerase II-DSIF-SPT6-PAF1c-TFIIS-IWS1-nucleosome, bp +27 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Markert, J, Farnung, L. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription.
Science, 387, 2025
|
|
9EGX
 
 | | RNA polymerase II-DSIF-SPT6-PAF1c-TFIIS-IWS1-hexasome, bp +27 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Markert, J, Farnung, L. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription.
Science, 387, 2025
|
|
9EGZ
 
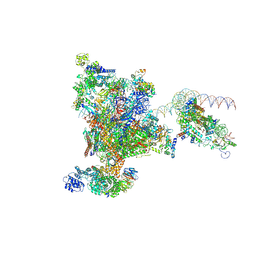 | |
9EH0
 
 | |
9EH2
 
 | |
9EAR
 
 | | CHD1-nucleosome complex (closed state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1, ... | | Authors: | James, A.M, Farnung, L. | | Deposit date: | 2024-11-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of human CHD1 nucleosome recruitment and pausing.
Mol.Cell, 85, 2025
|
|
9EH1
 
 | |
8P4B
 
 | | Structural insights into human co-transcriptional capping - structure 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*AP*GP*CP*GP*GP*CP*GP*GP*AP*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*AP*GP*CP*TP*G)-3'), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4C
 
 | | Structural insights into human co-transcriptional capping - structure 3 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4A
 
 | | Structural insights into human co-transcriptional capping - structure 1 | | Descriptor: | DNA (29-MER), DNA (38-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4D
 
 | | Structural insights into human co-transcriptional capping - structure 4 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
9GD0
 
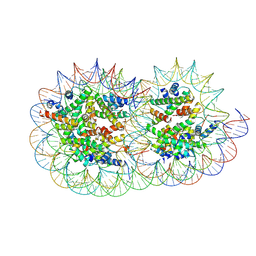 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD2
 
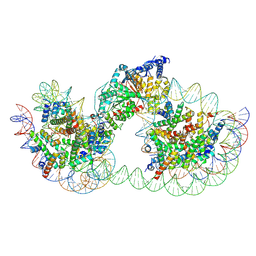 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD1
 
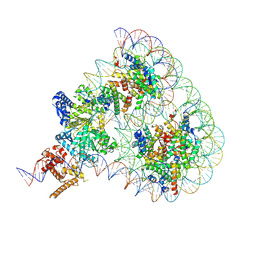 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD3
 
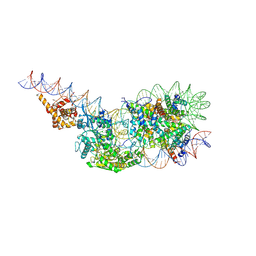 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
6S01
 
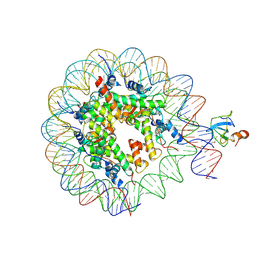 | | Structure of LEDGF PWWP domain bound H3K36 methylated nucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Wang, H, Farnung, L, Dienemann, C, Cramer, P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TED
 
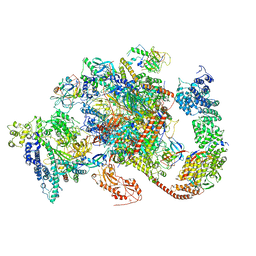 | | Structure of complete, activated transcription complex Pol II-DSIF-PAF-SPT6 uncovers allosteric elongation activation by RTF1 | | Descriptor: | DNA (37-MER), DNA-directed RNA polymerase II subunit RPB9, DNA-directed RNA polymerase subunit, ... | | Authors: | Vos, S.M, Farnung, L, Cramer, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
