6EMT
 
 | |
6EMV
 
 | |
6EMS
 
 | |
6EMU
 
 | |
1X83
 
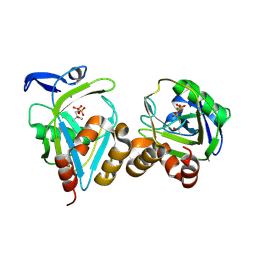 | | Y104F IPP isomerase reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
1X84
 
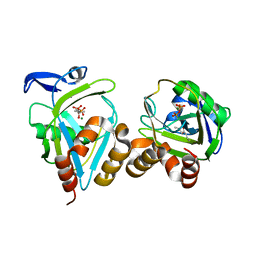 | | IPP isomerase (wt) reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
5NE9
 
 | |
5NE6
 
 | |
5NE8
 
 | |
5NE7
 
 | |
1OW2
 
 | |
1NFS
 
 | |
1NFZ
 
 | |
1PPW
 
 | |
1PPV
 
 | |
3LGA
 
 | |
3LHD
 
 | | Crystal structure of P. abyssi tRNA m1A58 methyltransferase in complex with S-adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, putative | | Authors: | Guelorget, A, Golinelli-Pimpaneau, B, Wouters, J, Barbey, C. | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase.
Nucleic Acids Res., 38, 2010
|
|
3MB5
 
 | |
