5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOS
 
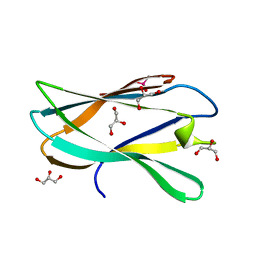 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4B1M
 
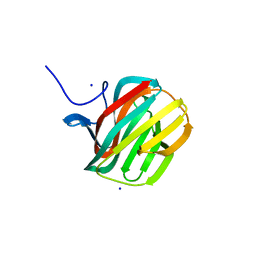 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B1L
 
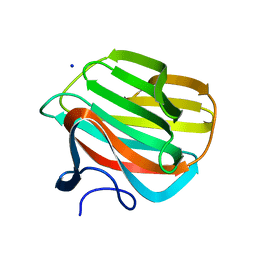 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1FOB
 
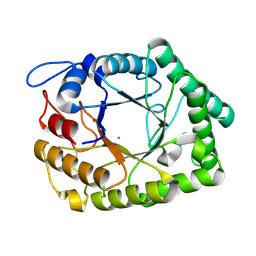 | |
1FHL
 
 | |
4AZZ
 
 | |
4D3L
 
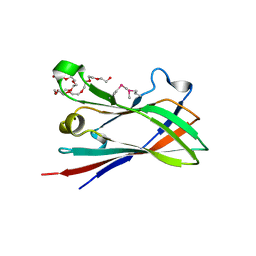 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 in the orthorhombic form | | Descriptor: | (3S)-3-HYDROXYHEPTANEDIOIC ACID, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-10-22 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2WZ8
 
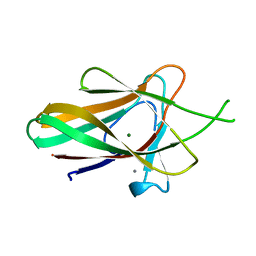 | | Family 35 carbohydrate binding module from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOME PROTEIN DOCKERIN TYPE I, MAGNESIUM ION | | Authors: | Gloster, T.M, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-11-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Signature Active Site Architectures Illuminate the Molecular Basis for Ligand Specificity in Family 35 Carbohydrate Binding Module .
Biochemistry, 49, 2010
|
|
2XN2
 
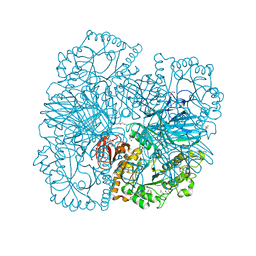 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN1
 
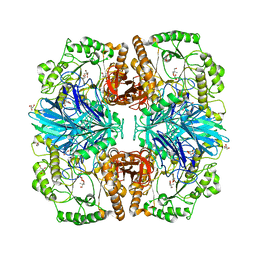 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN0
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, PLATINUM (II) ION | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
