7VLE
 
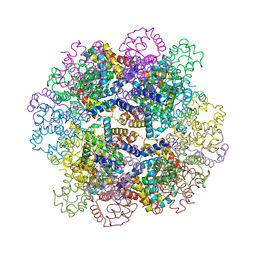 | | Oxy-deoxy intermediate of V2 hemoglobin at 55% oxygen saturation | | Descriptor: | Extracellular A1 globin, Extracellular A2 globin, Extracellular B1 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLD
 
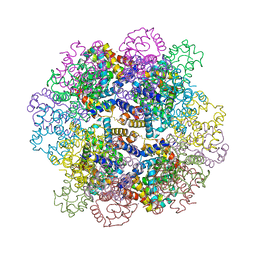 | | Oxy-deoxy intermediate of V2 hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLF
 
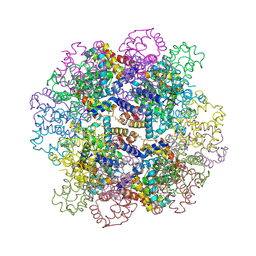 | | Oxy-deoxy intermediate of V2 hemoglobin at 26% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLC
 
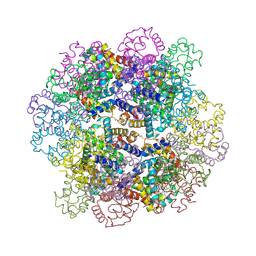 | | Oxy-deoxy intermediate of V2 hemoglobin at 78% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
4KEQ
 
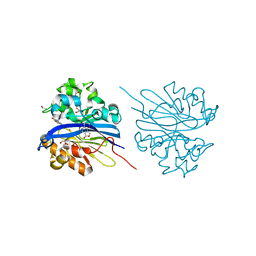 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8WO8
 
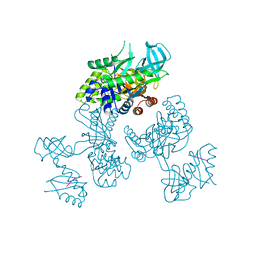 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
4KEP
 
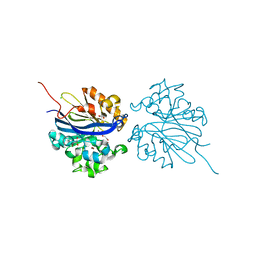 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5HNC
 
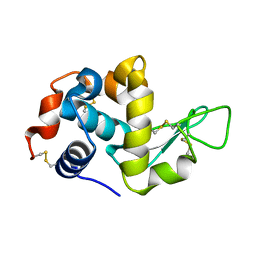 | | Synchrotron X-ray single crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Kusaka, K, Baba, S, Mizuno, N, Kimura, T. | | Deposit date: | 2016-01-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Neutron and X-ray single-crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5HNL
 
 | | In-house X-ray single crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Kusaka, K, Baba, S, Mizuno, N, Kimura, T. | | Deposit date: | 2016-01-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Neutron and X-ray single-crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
2III
 
 | | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5
To be Published
|
|
5B2Z
 
 | | H-Ras WT in complex with GppNHp (state 2*) before structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-07 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
2IDE
 
 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
2IIH
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
2IRP
 
 | | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Putative aldolase class 2 protein aq_1979 | | Authors: | Jeyakanthan, J, Gayathri, D, Yogavel, M, Velmurugan, D, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5
To be Published
|
|
2IEX
 
 | | Crystal structure of dihydroxynapthoic acid synthetase (GK2873) from Geobacillus kaustophilus HTA426 | | Descriptor: | Dihydroxynapthoic acid synthetase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, BaBa, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of dihydroxynapthoic acid synthetase (GK2873) from Geobacillus kaustophilus HTA426
To be Published
|
|
2IS8
 
 | | Crystal structure of the Molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8 | | Descriptor: | FORMIC ACID, Molybdopterin biosynthesis enzyme, MoaB | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8
To be Published
|
|
7QY4
 
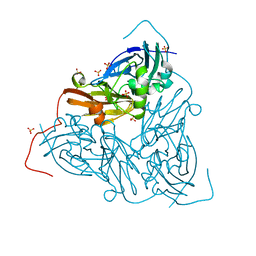 | | As isolated MSOX movie series dataset 5 (2 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QXK
 
 | | As isolated MSOX movie series dataset 1 (0.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QYC
 
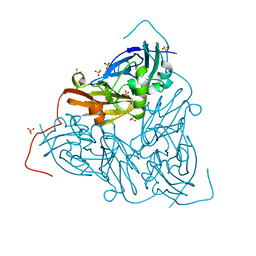 | | As isolated MSOX movie series dataset 20 (8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5B30
 
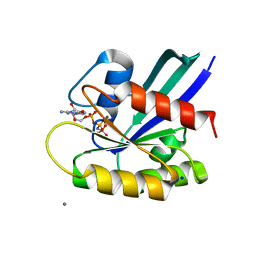 | | H-Ras WT in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
4XIG
 
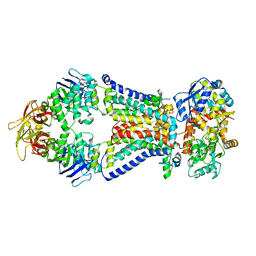 | | Crystal structure of bacterial alginate ABC transporter determined through humid air and glue-coating method | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
6L8S
 
 | | High resolution crystal structure of crustacean hemocyanin. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Masuda, T, Mikami, B, Baba, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Arch.Biochem.Biophys., 688, 2020
|
|
2FDT
 
 | | Solution structure of a conserved RNA hairpin of eel LINE UnaL2 | | Descriptor: | 36-mer | | Authors: | Nomura, Y, Baba, S, Nakazato, S, Sakamoto, T, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2
Nucleic Acids Res., 34, 2006
|
|
