2B4M
 
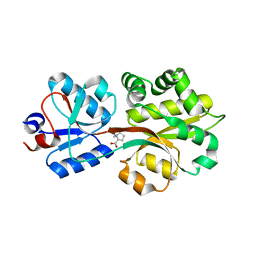 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2B4L
 
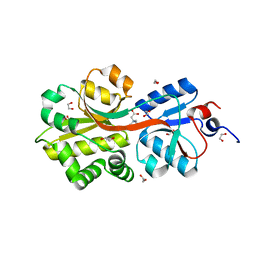 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2FCP
 
 | | FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) FROM E.COLI | | Descriptor: | 2-TRIDECANOYLOXY-PENTADECANOIC ACID, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Hofmann, E, Ferguson, A.D, Diederichs, K, Welte, W. | | Deposit date: | 1998-10-15 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide.
Science, 282, 1998
|
|
1PPR
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE | | Descriptor: | CHLOROPHYLL A, DIGALACTOSYL DIACYL GLYCEROL (DGDG), PERIDININ, ... | | Authors: | Hofmann, E, Welte, W, Diederichs, K. | | Deposit date: | 1996-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae.
Science, 272, 1996
|
|
5A3K
 
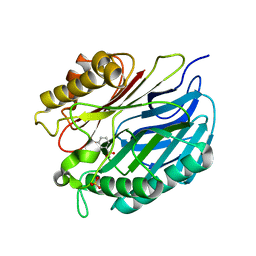 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
5AG3
 
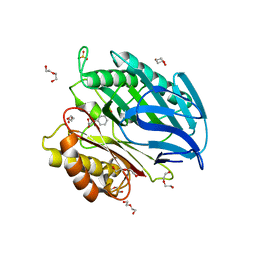 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-(2-CARBOXYETHYL)BENZOIC ACID, DI(HYDROXYETHYL)ETHER, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, ... | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-01-28 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
1A0S
 
 | | SUCROSE-SPECIFIC PORIN | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
1A0T
 
 | | SUCROSE-SPECIFIC PORIN, WITH BOUND SUCROSE MOLECULES | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
1U07
 
 | | Crystal Structure of the 92-residue C-term. part of TonB with significant structural changes compared to shorter fragments | | Descriptor: | TonB protein | | Authors: | Koedding, J, Killig, F, Polzer, P, Howard, S.P, Diederichs, K, Welte, W. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of a 92-residue c-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments
J.Biol.Chem., 280, 2005
|
|
1EU8
 
 | | STRUCTURE OF TREHALOSE MALTOSE BINDING PROTEIN FROM THERMOCOCCUS LITORALIS | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, TREHALOSE/MALTOSE BINDING PROTEIN, ... | | Authors: | Diez, J, Diederichs, K, Greller, G, Horlacher, R, Boos, W, Welte, W. | | Deposit date: | 2000-04-14 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic Archaeon Thermococcus litoralis at 1.85 A.
J.Mol.Biol., 305, 2001
|
|
1OH2
 
 | |
1QXX
 
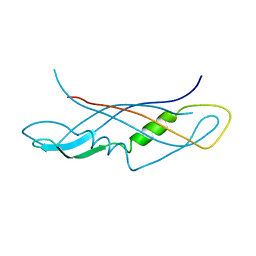 | | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF TONB | | Descriptor: | TonB protein | | Authors: | Koedding, J, Howard, P, Kaufmann, L, Polzer, P, Lustig, A, Welte, W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli.
J.Biol.Chem., 279, 2004
|
|
1QZ1
 
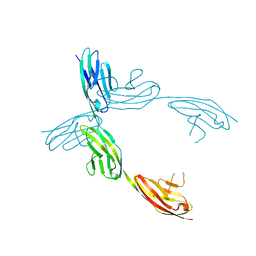 | | Crystal Structure of the Ig 1-2-3 fragment of NCAM | | Descriptor: | Neural cell adhesion molecule 1, 140 kDa isoform | | Authors: | Soroka, V, Kolkova, K, Kastrup, J.S, Diederichs, K, Breed, J, Kiselyov, V.V, Poulsen, F.M, Larsen, I.K, Welte, W, Berezin, V, Bock, E, Kasper, C. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion
Structure, 11, 2003
|
|
3QPH
 
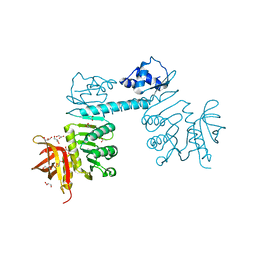 | | The three-dimensional structure of TrmB, a global transcriptional regulator of the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose | | Descriptor: | ACETATE ION, GLYCEROL, TrmB, ... | | Authors: | Krug, M, Lee, S.-J, Boos, W, Welte, W, Diederichs, K. | | Deposit date: | 2011-02-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | The three-dimensional structure of TrmB, a transcriptional regulator of dual function in the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose.
Protein Sci., 22, 2013
|
|
1R9Q
 
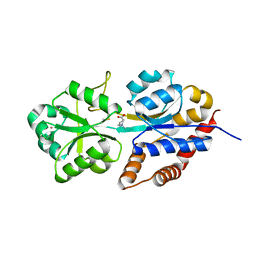 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
1R9L
 
 | | structure analysis of ProX in complex with glycine betaine | | Descriptor: | Glycine betaine-binding periplasmic protein, TRIMETHYL GLYCINE, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
1RYI
 
 | | STRUCTURE OF GLYCINE OXIDASE WITH BOUND INHIBITOR GLYCOLATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE OXIDASE, GLYCOLIC ACID | | Authors: | Moertl, M, Diederichs, K, Welte, W, Pollegioni, L, Molla, G, Motteran, L, Andriolo, G, Pilone, M.S. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function correlation in glycine oxidase from Bacillus subtilis
J.Biol.Chem., 279, 2004
|
|
1SW2
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with glycine betaine | | Descriptor: | TRIMETHYL GLYCINE, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1URS
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1URD
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius provide insight into acid stability of proteins | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1SW4
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with trimethyl ammonium | | Descriptor: | CHLORIDE ION, TETRAMETHYLAMMONIUM ION, ZINC ION, ... | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1SW1
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, ZINC ION, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1URG
 
 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1SW5
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in the ligand free form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1T92
 
 | | Crystal structure of N-terminal truncated pseudopilin PulG | | Descriptor: | General secretion pathway protein G, ZINC ION | | Authors: | Koehler, R, Schaefer, K, Mueller, S, Vignon, G, Diederichs, K, Philippsen, A, Ringler, P, Pugsley, A.P, Engel, A, Welte, W. | | Deposit date: | 2004-05-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and assembly of the pseudopilin PulG.
Mol.Microbiol., 54, 2004
|
|
