2ZVB
 
 | |
2DSJ
 
 | |
2DVN
 
 | |
2DVL
 
 | |
2DY0
 
 | |
2E18
 
 | |
2EKQ
 
 | |
2E9Y
 
 | |
3AQA
 
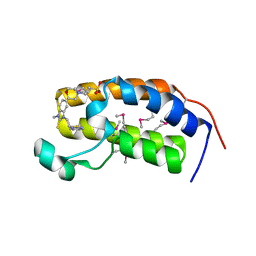 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
2ENW
 
 | |
2EJJ
 
 | |
2E4R
 
 | |
2ELD
 
 | |
2EJK
 
 | |
2EKL
 
 | |
2E37
 
 | |
2EKM
 
 | |
2ELC
 
 | |
2ZV3
 
 | |
2EKN
 
 | |
2EMU
 
 | |
2ZVC
 
 | |
2EMR
 
 | |
2EN5
 
 | |
2E4N
 
 | |
