7EEV
 
 | | Structure of UTP cyclohydrolase | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, GTP cyclohydrolase II, ZINC ION | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2022-09-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
7EJ3
 
 | | UTP cyclohydrolase | | Descriptor: | DIPHOSPHATE, GLYCINE, GTP cyclohydrolase II, ... | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-04-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
8ID7
 
 | |
8ID0
 
 | | Crystal structure of PflD bound to 1,5-anhydromannitol-6-phosphate in Streptococcus dysgalactiae subsp. equisimilis | | Descriptor: | [(2R,3S,4R,5R)-3,4,5-tris(oxidanyl)oxan-2-yl]methyl dihydrogen phosphate, formate C-acetyltransferase | | Authors: | Ma, K.L, Zhang, Y. | | Deposit date: | 2023-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
7VUA
 
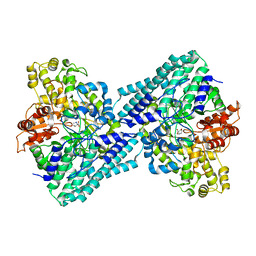 | | Anaerobic hydroxyproline degradation involving C-N cleavage by a glycyl radical enzyme | | Descriptor: | (4S)-4-hydroxy-D-proline, HplG | | Authors: | Duan, Y, Lu, Q, Yuchi, Z, Zhang, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Anaerobic Hydroxyproline Degradation Involving C-N Cleavage by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
8YHX
 
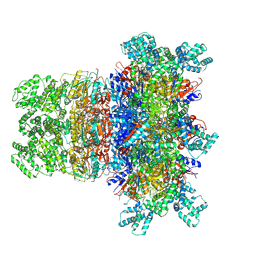 | | Cryo-EM structure of the trimeric HerA | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, DUF87 domain-containing protein, SIR2 family protein | | Authors: | Zhen, X, Zhou, B, Xiong, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Mechanistic basis for the allosteric activation of NADase activity in the Sir2-HerA antiphage defense system.
Nat Commun, 15, 2024
|
|
8YHO
 
 | | Cryo-EM structure of the trimeric HerA | | Descriptor: | DUF87 domain-containing protein | | Authors: | Zhen, X, Xiong, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanistic basis for the allosteric activation of NADase activity in the Sir2-HerA antiphage defense system.
Nat Commun, 15, 2024
|
|
8Z0G
 
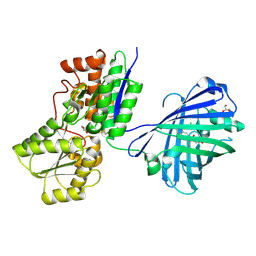 | | Crystal structure of NeIle complexed with isoleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Gabdukhakov, A, Subach, F.V. | | Deposit date: | 2024-04-09 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NeIle, a Genetically Encoded Indicator for Branched-Chain Amino Acids Based on mNeonGreen Fluorescent Protein and LIVBP Protein.
ACS Sens, 9, 2024
|
|
8YG6
 
 | | The pre-fusion structure of baculovirus fusion protein GP64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major envelope glycoprotein | | Authors: | Du, D, Guo, J, Li, S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch.
Nat Commun, 15, 2024
|
|
8YG8
 
 | | The early intermediate structure of baculovirus fusion protein GP64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major envelope glycoprotein | | Authors: | Du, D, Guo, J, Li, S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch.
Nat Commun, 15, 2024
|
|
8YJO
 
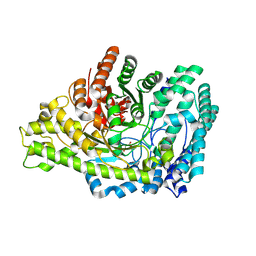 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJN
 
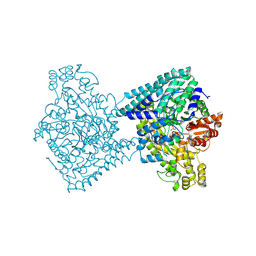 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
7E7L
 
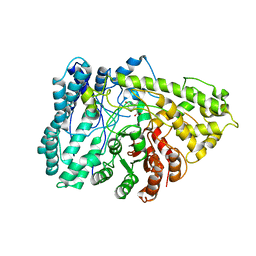 | | The crystal structure of arylacetate decarboxylase from Olsenella scatoligenes. | | Descriptor: | 4-HYDROXYPHENYLACETATE, Hydroxyphenylacetic acid decarboxylase | | Authors: | Lu, Q, Duan, Y, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The Glycyl Radical Enzyme Arylacetate Decarboxylase from Olsenella scatoligenes
Acs Catalysis, 11, 2021
|
|
7E39
 
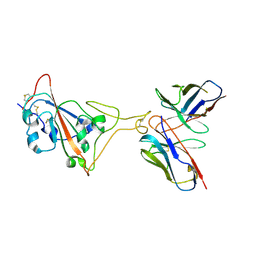 | | SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (State 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Ab4, Light Chain of Ab4, ... | | Authors: | Liu, C. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants.
Cell Discov, 7, 2021
|
|
7E3B
 
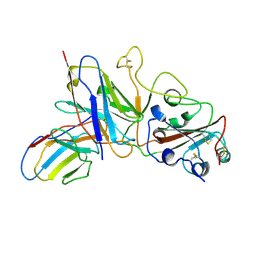 | |
7E3C
 
 | |
