7MA5
 
 | | HIV-1 Protease (I84V) in Complex with UMass4 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzodioxol-5-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass4
To Be Published
|
|
7MA3
 
 | | HIV-1 Protease (I84V) in Complex with UMass2 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}propyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass2
To Be Published
|
|
7MA4
 
 | | HIV-1 Protease (I84V) in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass3
To Be Published
|
|
7L0D
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML188 | | Descriptor: | 3C-like proteinase, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Lockbaum, G.J, Lee, J.M, Reyes, A.C, Nalivaika, E.A, Ali, A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188.
Viruses, 13, 2021
|
|
7MM6
 
 | |
7MMJ
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-08 | | Descriptor: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM7
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-23 | | Descriptor: | (2S)-1,1,1-trifluoropropan-2-yl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM5
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-60 | | Descriptor: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMG
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-58 | | Descriptor: | 1-(trifluoromethyl)cyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, ARGININE, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMH
 
 | |
7MM2
 
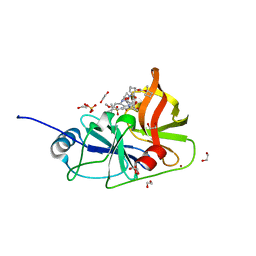 | | Crystal structure of HCV NS3/4A protease in complex with NR02-61 | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4a protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMF
 
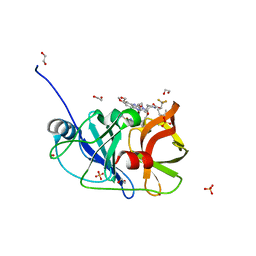 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-60 | | Descriptor: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMI
 
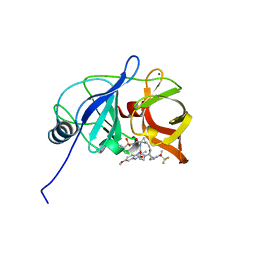 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-23 | | Descriptor: | (2S)-1,1,1-trifluoropropan-2-yl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3/4A protease, ZINC ION | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MML
 
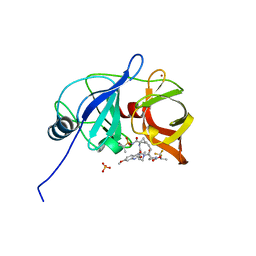 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-145 | | Descriptor: | (2R)-1,1,1-trifluoropropan-2-yl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM9
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-149 | | Descriptor: | 1,2-ETHANEDIOL, 3,3-difluorocyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMK
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-149 | | Descriptor: | 1,2-ETHANEDIOL, 3,3-difluorocyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMA
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-145 | | Descriptor: | (2R)-1,1,1-trifluoropropan-2-yl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM4
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM3
 
 | |
7MMB
 
 | |
7MMC
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7R8O
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C548 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8N
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C051 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8L
 
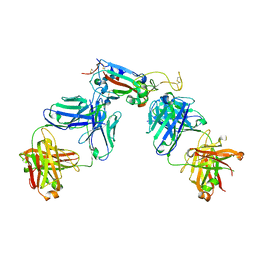 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody C099 and CR3022 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain, CR3022 Fab heavy chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8M
 
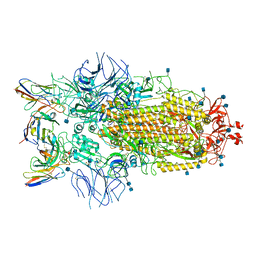 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C032 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
