6Q0S
 
 | |
6PWQ
 
 | | Crystal structure of Levansucrase from Bacillus subtilis mutant S164A at 2.6 A | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase family 68 protein, ... | | Authors: | Diaz-Vilchis, A, Rodriguez-Alegria, M.E, Ortiz-Soto, M.E, Rudino-Pinera, E, Lopez-Munguia, A. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Implications of the mutation S164A on Bacillus subtilis levansucrase product specificity and insights into protein interactions acting upon levan synthesis.
Int.J.Biol.Macromol., 161, 2020
|
|
6PYC
 
 | | Structure of kappa-on-heavy (KoH) antibody Fab bound to the cardiac hormone marinobufagenin | | Descriptor: | (3beta,5beta,14alpha,15beta)-3,5-dihydroxy-14,15-epoxybufa-20,22-dienolide, KoH body Fab heavy chain, KoH body Fab light chain, ... | | Authors: | Franklin, M.C, Macdonald, L.E, McWhirter, J, Murphy, A.J. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Kappa-on-Heavy (KoH) bodies are a distinct class of fully-human antibody-like therapeutic agents with antigen-binding properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PU7
 
 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
6PYP
 
 | | Binary Complex of Human Glycerol 3-Phosphate Dehydrogenase, R269A mutant | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic, ... | | Authors: | Gulick, A.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modeling the Role of a Flexible Loop and Active Site Side Chains in Hydride Transfer Catalyzed by Glycerol-3-phosphate Dehydrogenase.
Acs Catalysis, 10, 2020
|
|
6QRQ
 
 | | Apo conformation of chemotaxis sensor ODP | | Descriptor: | Oxygen-binding diiron protein | | Authors: | Muok, A.R, Crane, B.R. | | Deposit date: | 2019-02-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QNM
 
 | |
6QWO
 
 | | Zinc-reconstituted ODP from T. maritima | | Descriptor: | Oxygen-binding diiron protein, ZINC ION | | Authors: | Muok, A.R, Crane, B.R. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QZY
 
 | | full length OphA V406P in complex with SAH | | Descriptor: | ASN-GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-PRO-ILE-GLY, MAGNESIUM ION, Peptide N-methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6QZZ
 
 | | full length OphA V404E in complex with SAH | | Descriptor: | Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6R00
 
 | | OphA DeltaC6 V404F complex with SAH | | Descriptor: | PHE-PRO-TRP-MVA-ILE-MVA-PHE-GLY-VAL-ILE-GLY-VAL-ILE-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6E7E
 
 | |
6EP9
 
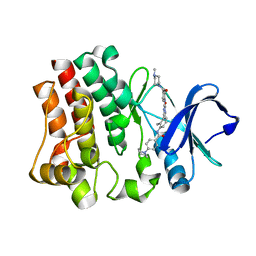 | |
6EQA
 
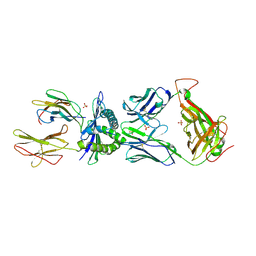 | | HLA class I histocompatibility antigen | | Descriptor: | ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | TCR-induced alteration of primary MHC peptide anchor residue.
Eur.J.Immunol., 49, 2019
|
|
6EQT
 
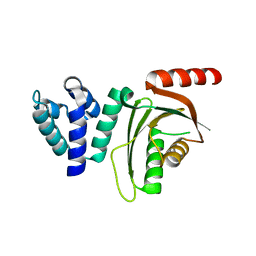 | |
6EE0
 
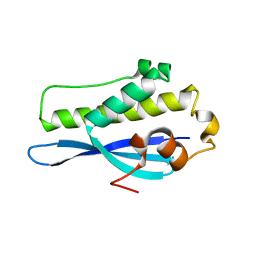 | | Crystal Structure of SNX23 PX domain | | Descriptor: | Kinesin-like protein KIF16B | | Authors: | Chandra, M, Collins, B.M. | | Deposit date: | 2018-08-12 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities.
Nat Commun, 10, 2019
|
|
6ECV
 
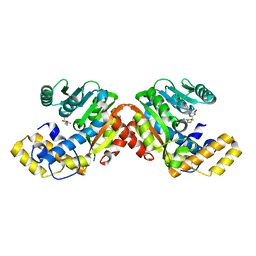 | | StiD O-MT residues 976-1266 | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECT
 
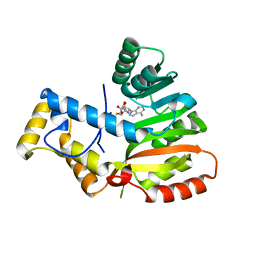 | | StiE O-MT residues 961-1257 | | Descriptor: | S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6E8E
 
 | |
6E8D
 
 | |
6E6A
 
 | | Triclinic crystal form of IncA G144A point mutant | | Descriptor: | Inclusion membrane protein A, SODIUM ION | | Authors: | Cingolani, G, Paumet, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA.
Nat Commun, 10, 2019
|
|
6ECM
 
 | |
6E8Y
 
 | | Unliganded Human Glycerol 3-Phosphate Dehydrogenase | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic, ... | | Authors: | Mydy, L.S, Gulick, A.M. | | Deposit date: | 2018-07-31 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human Glycerol 3-Phosphate Dehydrogenase: X-ray Crystal Structures That Guide the Interpretation of Mutagenesis Studies.
Biochemistry, 58, 2019
|
|
6E3C
 
 | |
6E90
 
 | | Ternary complex of human glycerol 3-phosphate dehydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, CALCIUM ION, ... | | Authors: | Mydy, L.S, Gulick, A.M. | | Deposit date: | 2018-07-31 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human Glycerol 3-Phosphate Dehydrogenase: X-ray Crystal Structures That Guide the Interpretation of Mutagenesis Studies.
Biochemistry, 58, 2019
|
|
