1U6I
 
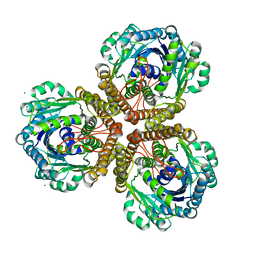 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1KIQ
 
 | |
1KIR
 
 | |
1KIP
 
 | |
1OGY
 
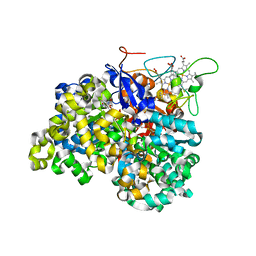 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | Authors: | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | Deposit date: | 2003-05-19 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
1JCK
 
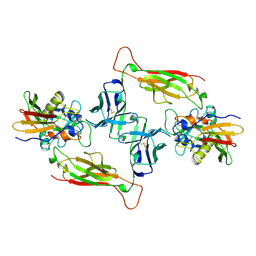 | |
7GS3
 
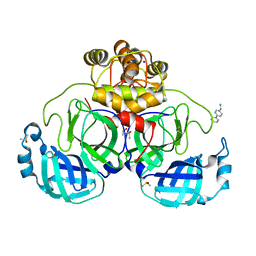 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-26 | | Descriptor: | (6-phenylpyridin-3-yl)methanamine, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRZ
 
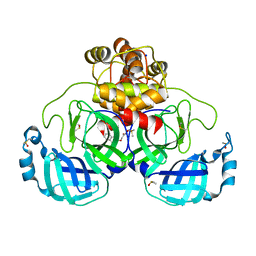 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-22 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N,N-dimethyl-2-[(naphthalen-2-yl)oxy]acetamide, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-28 | | Descriptor: | (3R)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS2
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-25 | | Descriptor: | 3-(pyridin-3-yl)benzoic acid, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-4 | | Descriptor: | 3C-like proteinase nsp5, 5-chloropyridin-3-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS6
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-29 | | Descriptor: | (3S)-4,7-dichloro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRP
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-12 | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)methanamine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRG
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-3 | | Descriptor: | 3,5-dichloropyridin-4-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-7 | | Descriptor: | (6-fluoro-2H,4H-1,3-benzodioxin-8-yl)methanol, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRT
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-16 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2,3-dihydro-1-benzofuran-5-yl)methyl]benzamide, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-14 | | Descriptor: | 3C-like proteinase nsp5, 5-(3-cyclohexylprop-1-yn-1-yl)pyridine-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRI
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-5 | | Descriptor: | (1S)-1-(1H-pyrazol-5-yl)ethan-1-ol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-21 | | Descriptor: | 1-(3,5-dichlorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRL
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-8 | | Descriptor: | 3C-like proteinase nsp5, 4-(4,5-dibromo-2H-1,2,3-triazol-2-yl)butan-2-one, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS4
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-27 | | Descriptor: | 3C-like proteinase nsp5, 7-(hydroxymethyl)-3-methyl-6~{H}-[1,3]thiazolo[3,2-a]pyrimidin-5-one, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRO
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-11 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRE
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-1 | | Descriptor: | 3C-like proteinase nsp5, 4-[3-(trifluoromethyl)-1H-pyrazol-5-yl]pyridine, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A.M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRQ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-13 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRM
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-9 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
