3FUH
 
 | |
3FTX
 
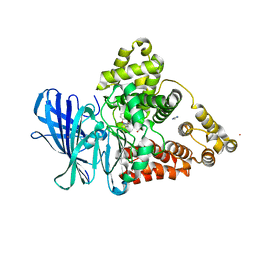
 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
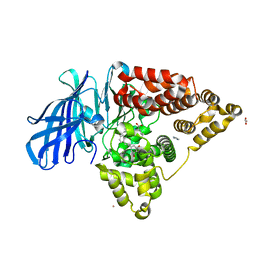
3FUI
 
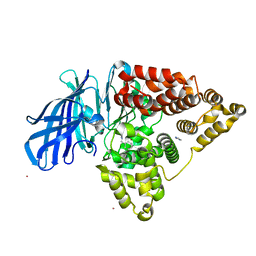 | |
3FU5
 
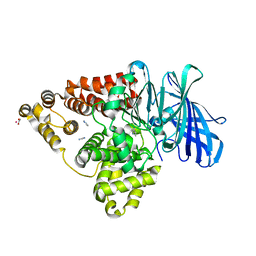 | |
3FUJ
 
 | |
5X1U
 
 | |
3FTY
 
 | |
3FUF
 
 | |
3FTW
 
 | |
3FUD
 
 | |
3FUE
 
 | |
3FDZ
 
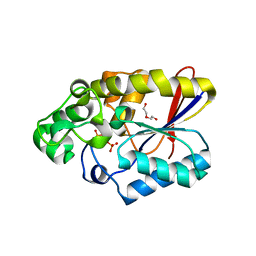 | | Crystal structure of phosphoglyceromutase from burkholderia pseudomallei 1710b with bound 2,3-diphosphoglyceric acid and 3-phosphoglyceric acid | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3GW8
 
 | |
3GP5
 
 | |
3FTV
 
 | |
3FUM
 
 | |
3WO5
 
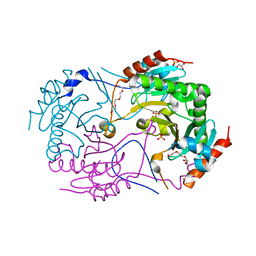 | | Crystal structure of S147Q of Rv2613c from Mycobacterium tuberculosis | | Descriptor: | AP-4-A phosphorylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mori, S, Wachino, J, Arakawa, Y, Shibayama, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Role of Ser-147 and Ala-149 in catalytic activity of diadenosine tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
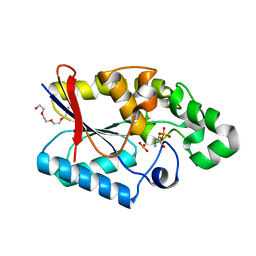
3GP3
 
 | |
5X42
 
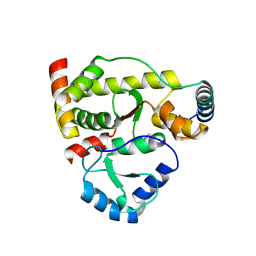 | | Structure of DotL(590-659)-DotN derived from Legionella pneumophila | | Descriptor: | DotN, IcmJ (DotN), IcmO (DotL), ... | | Authors: | Kwak, M.J, Kim, J.D, Oh, B.H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
2MUQ
 
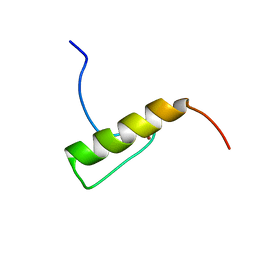 | | Solution Structure of the Human FAAP20 UBZ | | Descriptor: | Fanconi anemia-associated protein of 20 kDa, ZINC ION | | Authors: | Wojtaszek, J.L, Wang, S, Zhou, P. | | Deposit date: | 2014-09-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain.
Nucleic Acids Res., 42, 2014
|
|
2MUR
 
 | |
5X1H
 
 | | Structure of Legionella pneumophila DotN | | Descriptor: | IcmJ (DotN), ZINC ION | | Authors: | Kwak, M.J, Oh, B.H. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
2KX2
 
 | |
5YSS
 
 | |
2Q8A
 
 | | Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody | | Descriptor: | 1F9 heavy chain, 1F9 light chain, Apical membrane antigen 1 | | Authors: | Gupta, A, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2007-06-10 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody.
PLoS Pathog., 3, 2007
|
|
