4ZHJ
 
 | |
5B6L
 
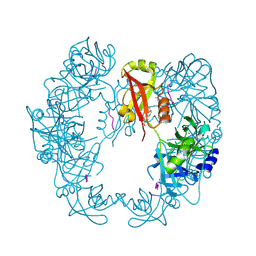 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
5CHE
 
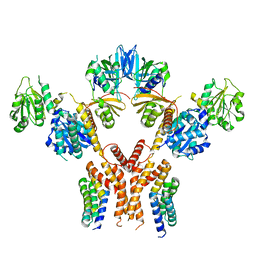 | |
7BOR
 
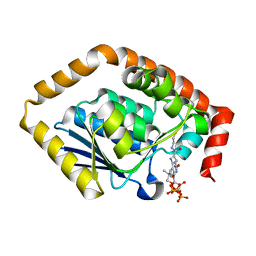 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7CRD
 
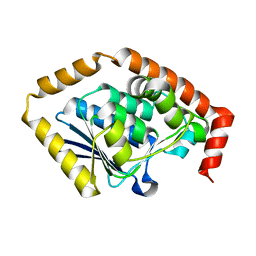 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7F4S
 
 | | Crystal structure of TthMTA1-PteMTA9 complex | | Descriptor: | MT-a70 family protein, MTA9 | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4N
 
 | | Crystal structure of SAH-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4Q
 
 | | Crystal structure of SAH-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4O
 
 | | Crystal structure of MTA1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4P
 
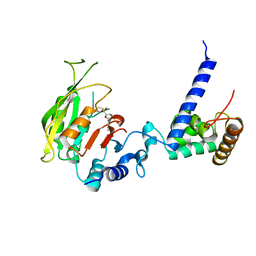 | | Crystal structure of SAM-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4M
 
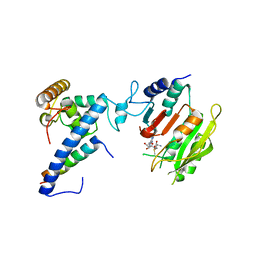 | | Crystal structure of SAM-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4T
 
 | |
7F4L
 
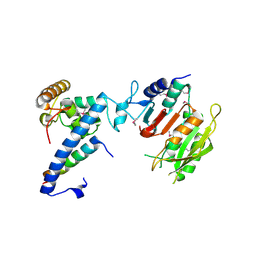 | | Crystal structure of MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
8IYA
 
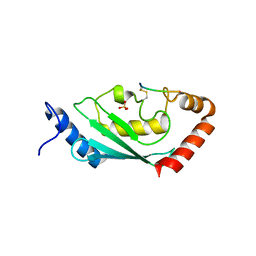 | | Complex of SETDB1-derived peptide bound to UBE2E1 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, SULFATE ION, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Du, Y.X, Liu, L. | | Deposit date: | 2023-04-04 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complex of SETDB1-derived peptide bound to UBE2E1
To Be Published
|
|
8J1P
 
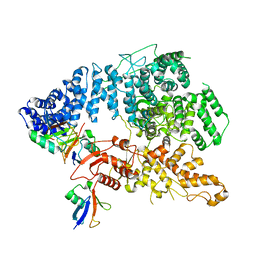 | | Cryo-EM structure of Ufd4 in complex with K29/48 triUb | | Descriptor: | Ubiquitin, Ubiquitin fusion degradation protein 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural Insights into the Molecular Mechanism of Ufd4-catalyzed Elongation of K48-linked Ubiquitin Chain through Lys29 Linkage
To Be Published
|
|
8J1R
 
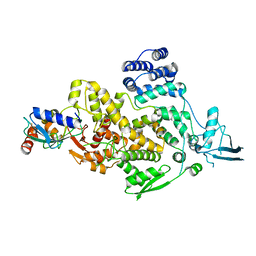 | | cryo-EM structures of Ufd4 in complex with Ubc4-Ub | | Descriptor: | Ubiquitin fusion degradation protein 4, Ubiquitin-conjugating enzyme E2 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Cai, H.Y, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Visualization of HECT-E3 Ufd4 accepting and transferring Ubiquitin to Form K29/K48-branched Polyubiquitination on N-degron. bioRxiv,doi: ttps://doi.org/10.1101/2023.05.23.542033
To Be Published
|
|
7YR7
 
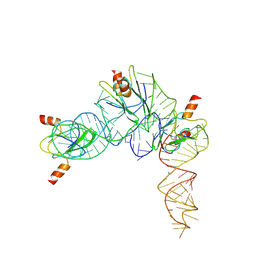 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
5DQR
 
 | |
5FEJ
 
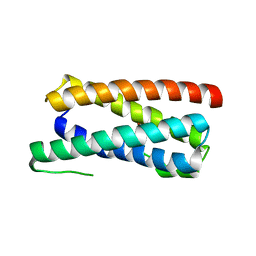 | | CopM in the Cu(I)-bound form | | Descriptor: | COPPER (I) ION, CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-17 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFA
 
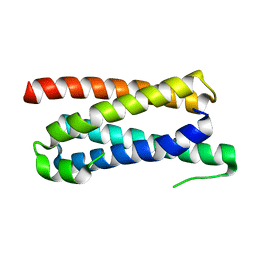 | |
5FFC
 
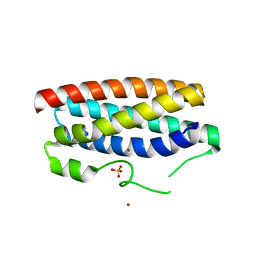 | | CopM in the Cu(II)-bound form | | Descriptor: | COPPER (II) ION, CopM, SULFATE ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFE
 
 | | CopM in the Ag-bound form (by soaking) | | Descriptor: | CopM, SILVER ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFB
 
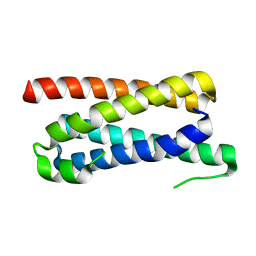 | | CopM in the apo form | | Descriptor: | CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFD
 
 | |
5FW5
 
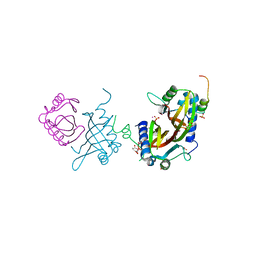 | | Crystal structure of human G3BP1 in complex with Semliki Forest Virus nsP3-25 comprising two FGDF motives | | Descriptor: | ACETATE ION, GLYCEROL, NON-STRUCTURAL PROTEIN 3, ... | | Authors: | Schulte, T, Liu, L, Panas, M.D, Thaa, B, Goette, B, Achour, A, McInerney, G.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.
Open Biol, 6, 2016
|
|
