4W6U
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115H/T118H With Nickel Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, NICKEL (II) ION, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W77
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 3 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7D
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26H With Copper Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6T
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115H/T118H With Copper Mediated Crystal Contacts, P 43 21 2 Space Group | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, COPPER (II) ION, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W76
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 2 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7E
 
 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H With Copper Mediated Crystal Contacts, P 41 21 2 Space Group | | Descriptor: | COPPER (II) ION, IMIDAZOLE, fluorescent protein D21H/K26H | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7R
 
 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H Copper Mediated Dimer, P 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6B
 
 | | Crystal Structure of a Superfolder GFP Mutant K26C Disulfide Dimer, P 21 21 21 Space Group | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, fluorescent protein K26C | | Authors: | Pashkov, I, Sawaya, M.R, Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6G
 
 | |
4W6J
 
 | | Crystal Structure of Full-Length Split GFP Mutant D117C Disulfide Dimer, P 31 2 1 Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, fluorescent protein D117C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6O
 
 | |
4W6R
 
 | |
4W6N
 
 | |
4W72
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115C/T118H Disulfide Dimer With Copper Mediated Crystal Contacts, P 21 21 21, Form 1 | | Descriptor: | COPPER (II) ION, fluorescent protein E115C/T118H | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7C
 
 | |
4W7X
 
 | |
6NKQ
 
 | |
1AIJ
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-18 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
1AIG
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE D+QB-CHARGE SEPARATED STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
1PSS
 
 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
1PST
 
 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
5VL4
 
 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
1PVC
 
 | |
1F1O
 
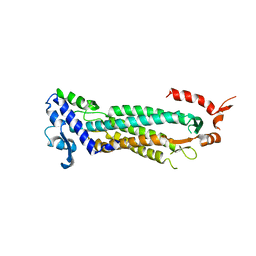 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
2FGY
 
 | |
