5TFU
 
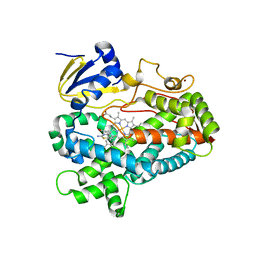 | | Structure of cytochrome P450 2D6 (CYP2D6) BACE1 inhibitor complex | | Descriptor: | (4S,6R)-4-[2,4-difluoro-5-({[1-(trifluoromethyl)cyclopropyl]amino}methyl)phenyl]-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2016-09-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality.
J. Med. Chem., 60, 2017
|
|
1K8K
 
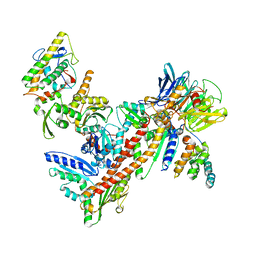 | | Crystal Structure of Arp2/3 Complex | | Descriptor: | ACTIN-LIKE PROTEIN 2, ACTIN-LIKE PROTEIN 3, ARP2/3 COMPLEX 16 KDA SUBUNIT, ... | | Authors: | Robinson, R.C, Turbedsky, K, Kaiser, D.A, Higgs, H.N, Marchand, J.-B, Choe, S, Pollard, T.D. | | Deposit date: | 2001-10-24 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Arp2/3 Complex
Science, 294, 2001
|
|
8QX8
 
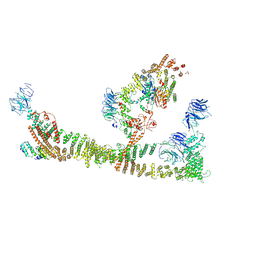 | | Endosomal membrane tethering complex CORVET | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar protein sorting-associated protein 16, ... | | Authors: | Shvarev, D, Ungermann, C, Moeller, A. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
4JT9
 
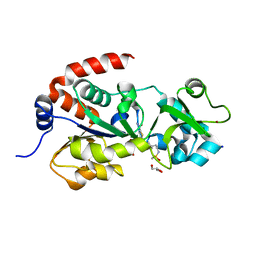 | | Crystal Structure of human SIRT3 with ELT inhibitor 3 [14-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide] | | Descriptor: | 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, GLYCEROL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
5UKP
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.1 Fab | | Descriptor: | DH522.1 Fab fragment heavy chain, DH522.1 Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
5UKN
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522UCA Fab | | Descriptor: | CHLORIDE ION, DH522UCA Fab fragment heavy chain, DH522UCA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
5UKR
 
 | |
5UKO
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522IA Fab | | Descriptor: | DH522IA Fab fragment heavy chain, DH522IA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
7A5X
 
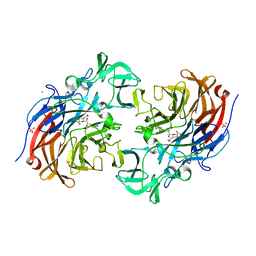 | | Two copies of the catalytic domain of NanA sialidase from Streptococcus pneumoniae juxtaposed in the P212121 space group, in complex with DANA derivatized with a PEG linker on the glycerol group. | | Descriptor: | (4~{S},5~{R},6~{R})-5-acetamido-6-[(1~{R},2~{S})-3-[[1-(2-ethoxyethyl)-1,2,3-triazol-4-yl]methylsulfanyl]-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bridot, C, Bouckaert, J. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polyvalent Transition-State Analogues of Sialyl Substrates Strongly Inhibit Bacterial Sialidases*.
Chemistry, 27, 2021
|
|
7QJT
 
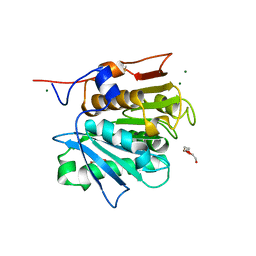 | | Crystal structure of a cutinase enzyme from Thermobifida cellulosilytica TB100 (711) | | Descriptor: | GLYCEROL, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
5UKQ
 
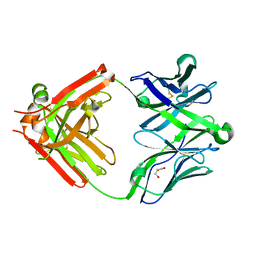 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.2 Fab | | Descriptor: | DH522.2 Fab fragment heavy chain, DH522.2 Fab fragment light chain, GLYCEROL | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
7QJM
 
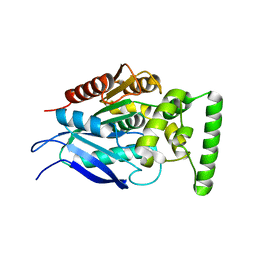 | |
7QJO
 
 | |
7QJN
 
 | | Crystal structure of an alpha/beta-hydrolase enzyme from Candidatus Kryptobacter tengchongensis (306) | | Descriptor: | Dienelactone hydrolase, PHOSPHATE ION | | Authors: | Zahn, M, Gill, R.S, Erickson, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJS
 
 | | Crystal structure of a cutinase enzyme from Thermobifida fusca YX (705) | | Descriptor: | Cutinase 2, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
4JT8
 
 | | Crystal Structure of human SIRT3 with ELT inhibitor 28 [4-(4-{2-[(2,2-dimethylpropanoyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide[ | | Descriptor: | 4-(4-{2-[(2,2-dimethylpropanoyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
7BHD
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
2R9L
 
 | | Polymerase Domain from Mycobacterium tuberculosis Ligase D in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*DGP*DCP*DCP*DGP*DCP*DAP*DAP*DCP*DGP*DCP*DA)-3'), ... | | Authors: | Brissett, N.C, Fox, G.C, Pitcher, R.S, Doherty, A.J. | | Deposit date: | 2007-09-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a NHEJ polymerase-mediated DNA synaptic complex
Science, 318, 2007
|
|
4JSR
 
 | | Crystal Structure of human SIRT3 with ELT inhibitor 11c [N-{2-[1-(6-carbamoylthieno[3,2-d]pyrimidin-4-yl)piperidin-4-yl]ethyl}-N'-ethylthiophene-2,5-dicarboxamide] | | Descriptor: | N-{2-[1-(6-carbamoylthieno[3,2-d]pyrimidin-4-yl)piperidin-4-yl]ethyl}-N'-ethylthiophene-2,5-dicarboxamide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
2IRX
 
 | |
2IRU
 
 | |
2IRY
 
 | |
6U6O
 
 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH846 | | Descriptor: | DH846 Fab heavy chain, DH846 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
6U6M
 
 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH840.1 | | Descriptor: | DH840.1 Fab heavy chain, DH840.1 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
6MSC
 
 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3-methyl-1-(3-methylpyridin-4-yl)-3H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
